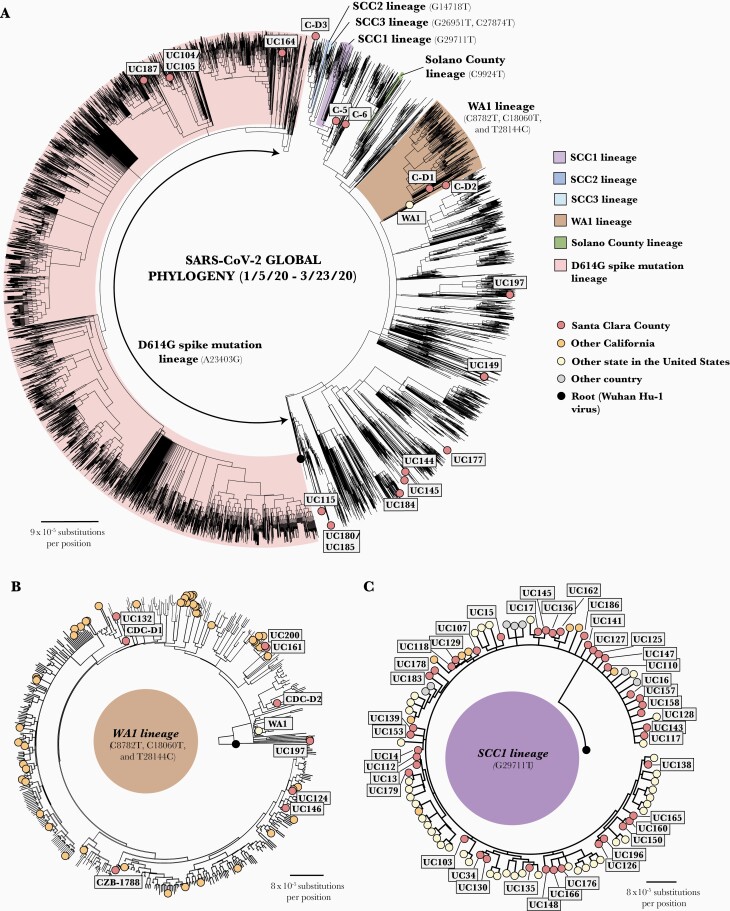
Figure 2.
Phylogenetic tree analysis of severe acute respiratory syndrome coronavirus (SARS-CoV-2) strains in Santa Clara county collected from 27 January to 21 March 2020. A, Radial view of Santa Clara SARS-CoV-2 viruses (n = 101) in the context of 19 922 global SARS-CoV-2 strains. Viral lineages are labeled on the tree and color coded. B, Radial view of WA1 lineage. The death case C-D2 (possessing 3 single-nucleotide variants [SNVs]) is situated on a side branch of the WA1 subtree, while the earlier death case C-D1 is positioned within the main WA1 cluster (with 5 common SNVs). Note that all genomes in the WA1 lineage are closely related to each other, and alignments can be artifactually driven by gaps (stretches of N’s) in the assembled genome. Thus, CZB-1788 is positioned away from C124 and UC146 despite all 3 individuals being passengers on the same cruise and having identical genomes. C, Overall view of SCC1 lineage sharing G29711T.