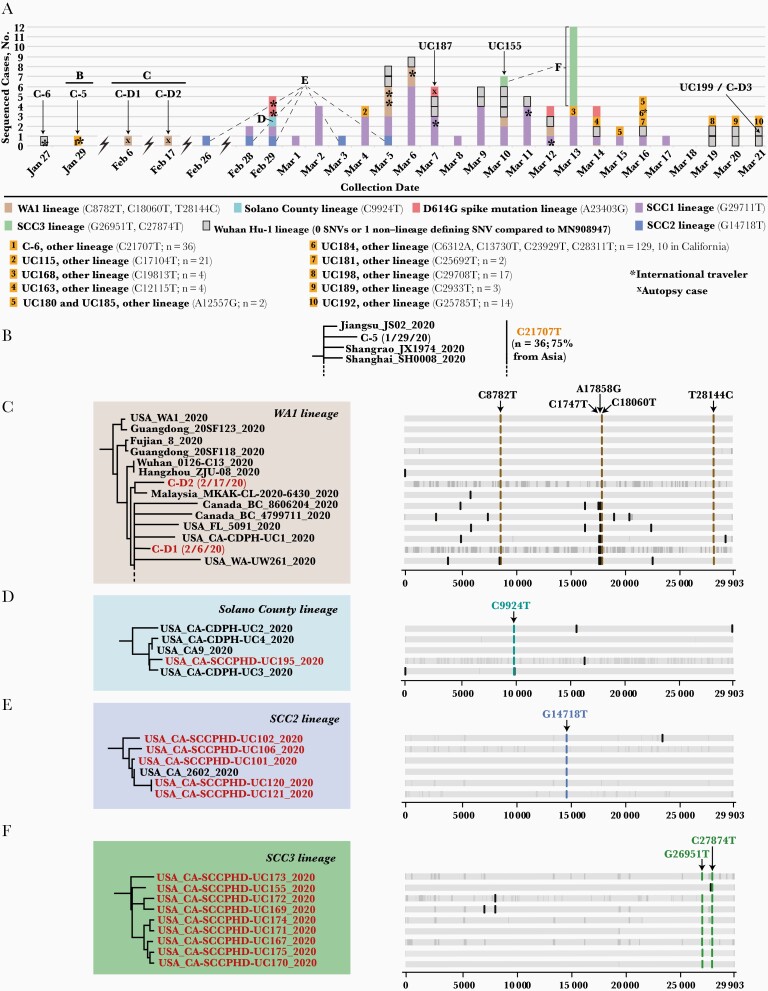
Figure 3.
Single-nucleotide variant (SNV) analysis of Santa Clara severe acute respiratory syndrome coronavirus 2 strains collected between 27 January and 21 March 2020. A, Daily composition of viral lineages in Santa Clara County. The 7 main lineages are represented in different colors, while the remaining 10 lineages are given numerical designations. B, C-5 and related strains from China. C, Phylogenetic tree of WA1 strains, including 2 early death cases and their defining SNVs. Note that given the size of the WA1 subtree, not all of the 9 WA1 genomes recovered in this study are shown (dashed line). D, Solano County lineage and defining SNV. E, SCC2 lineage and the defining G14718T. F, SCC3 lineage and defining G26951T and C27874 SNVs. In B–F, highlighted red font denotes the genome as belonging to the 101 cases in the study. In C–F, the vertical gray lines represent regions of the genome with missing coverage, represented as nucleotide stretches of N’s in the assembled sequence.