Abstract
During the 2015–16 winter, the US experienced a relatively mild influenza season compared to Taiwan, which had a higher number of total and severe cases. While H1N1pdm viruses dominated global surveillance efforts that season, the global distribution of genetic variants and their contributions to disease severity have not been investigated. Samples collected from influenza A-positive patients by the Johns Hopkins Center of Excellence for Influenza Research and Surveillance active surveillance in the emergency rooms in Baltimore, Maryland, USA, and northern Taiwan between November 2015 and April 2016, were processed for influenza A virus whole-genome sequencing. In Baltimore, the majority of the viruses were the H1N1pdm clade 6B.1 and no H1N1pdm clade 6B.2 viruses were detected. In northern Taiwan, more than half of the H1N1pdm viruses were clade 6B.1 and 38% were clade 6B.2, consistent with the global observation that most 6B.2 viruses circulated in Asia and not North America. Whole virus genome sequence analysis identified two genetic subgroups present in each of the 6B.1 and 6B.2 clades and one 6B.1 interclade reassortant virus. Clinical data showed 6B.2 patients had more disease symptoms including higher crude and inverse probability weighted odds of pneumonia than 6B.1 patients, suggesting 6B.2 circulation may be one of the reasons for the severe flu season in Taiwan. Local surveillance efforts linking H1N1pdm virus sequences to patient clinical and demographic data improve our understanding of influenza circulation and disease potential.
Keywords: influenza, H1N1pdm, whole-genome sequencing, disease severity, Taiwan, Baltimore
1. Introduction
The 2009 pandemic H1N1 influenza A virus (H1N1pdm) has supplanted the previous human H1N1 viruses to become the seasonal human H1N1 virus. In the 2015–16 influenza season, the northern hemisphere experienced its second global H1N1pdm-dominant year since 2009. The 2015–16 Influenza activity in the United States was considered moderate, with lower numbers of total cases and a later peak epidemic when compared with the previous three influenza seasons (Davlin et al. 2016). However, in Taiwan, the 2015–16 season was much more serious (Gong et al. 2018), with an earlier start and higher numbers of total as well as severe, influenza cases compared to the prior influenza season 2014–15 (Taiwan CDC 2016).
Influenza A viruses (IAV) are subdivided into subtypes based on the antibody response to the viral glycoproteins, hemagglutinin (HA) and neuraminidase (NA). Viruses are further divided into genetic clades based on HA sequences in order to monitor for mutations that might lead to antigenic drift. The 2015–16 northern hemisphere vaccine strain of H1N1pdm, A/California/07/2009, is defined as H1 clade 1. Prior to the 2015–16 season, H1N1pdm was dominated by clade 6B. The global rise of H1 clade 6B.1 carrying amino acid changes S84N, S162N and I216T from clade 6B was detected in human surveillance efforts starting in June 2015 and is now the dominant H1N1 clade. The H1 clade 6B.2 viruses (V152T, V173I of HA1 and E164G and D174E of HA2) emerged in July 2015, peaked in January 2016, were found primarily in Asia and disappeared from surveillance efforts at the end of 2016 (Neher and Bedford 2015; Bedford and Neher 2017). The majority of 6B.1 and 6B.2 viruses were antigenically similar to the recommended components of the 2015–16 Northern Hemisphere H1N1pdm influenza vaccine A/California/07/2009 (Chambers et al. 2016; Davlin et al. 2016; Komissarov et al. 2016; Taiwan CDC 2016), although antigenic difference between egg-adapted and circulating virus strains may have contributed to increased numbers of cases (Garretson et al. 2018).
The genome of IAV consists of eight segments of negative-sense RNA. In addition to HA mutations, mutations in the other seven viral gene segments also result in genetic variants which can alter virus fitness, subvert pre-existing immunity and/or reduce vaccine effectiveness. Any of these changes could potentially alter disease severity or total number of cases (Bouvier and Palese 2008). Next-generation sequencing provides a sensitive and rapid method for sequencing of viruses directly from human specimens without any need for virus isolation or culture—both of which are known to select for viruses bearing additional mutations (Zhou et al. 2009).
Most current global influenza surveillance systems lack connections between viral sequences and detailed patient demographic and clinical data. The Johns Hopkins Center of Excellence for Influenza Research and Surveillance (JH-CEIRS) has been actively performing human influenza surveillance since 2014. Surveillance efforts involve the connection of viral genome data and isolates with patient demographic and clinical data to gain more insights into how virus genetic variation can affect clinical disease. We hypothesized that sequence differences in influenza viruses could be associated with distinct clinical outcomes. This study involved concurrent, active influenza surveillance during the 2015–16 season in Baltimore, USA, and northern Taiwan to compare and contrast differential epidemics in the same year between the two geographic regions and to analyze possible disease severity with influenza viral genotypes.
2. Materials and methods
2.1 Active surveillance enrollment and sample collection
Institutional Review Boards at the Johns Hopkins University School of Medicine and Chang Gung Memorial Hospital provided ethical approval of the study (IRB00052743). From November 2015 to April 2016, active surveillance was performed in the adult emergency departments of the Johns Hopkins Medical Institutions (JHMI) at the East Baltimore and Bayview campuses in Baltimore, Maryland, USA, and Chang Gung Memorial Hospitals (CGMH) in the northern Taiwan metropolitan area, including hospitals in the Taipei, Linkou and Keelung Branches. Influenza-like illness was defined as documented or reported ≥38 °C fever and any of one of three respiratory symptoms including cough, headache and/or sore throat within the past seven days. Exclusion criteria included subjects who are unable to speak and understand English (in United States) or Mandarin (in Taiwan), unable to provide written informed consent, currently incarcerated or previously enrolled in the study during the same influenza season. Patients were approached by trained clinical coordinators who obtained written, informed consent before collected specimens and demographic and clinical data using a standard questionnaire. Data were confirmed by examination of the patient’s electronic health record. All data were de-identified and stored in a secure REDCap database (Harris et al. 2009; Harris et al. 2019).
2.2 Diagnosis and subtyping
Nasopharyngeal swabs or nasal washes of patients collected at the time of enrollment were tested for IAV infection using the Cepheid Xpert Flu/RSV test (Cepheid, Sunnyvale, CA). Samples that were IAV positive were further subtyped by quantitative, reverse transcription, real time polymerase chain reaction (qRT-PCR) with specific H1 or H3 primers and probes according to USA Centers for Disease Control and Prevention (CDC) protocols. Only H1N1pdm positive samples were moved on to whole genome sequencing (WGS) analysis.
2.3 Whole-genome sequencing
Viral RNA was isolated from the influenza A-positive clinical samples using the MagMax Viral RNA isolation reagent (Thermo Fisher) on an Eppendorf epMotion 5073 liquid handling workstation. Samples were processed for WGS using multi-segment PCR (Zhou et al. 2009) and then prepared for deep sequencing using the Nextera XT library preparation reagent (Illumina). Samples were sequenced on the Illumina NextSeq platform. Raw paired-end data were first processed through Trim galore! (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) with a quality score of 30 and an adapter sequence of CTGTCTCTTATACACATCT, only retaining pairs that both passed through this quality control step. The quality-trimmed reads were then aligned to the influenza vaccine strain reference sequence using bowtie2 (version 2.1.0) (Langmead and Salzberg 2012) with the ‘–very-sensitive-local’ option and converted to sorted BAM files using samtools (Li et al. 2009). Only sequences of all eight gene segments passed quality controls were included in the study. The sequences were submitted to NCBI GenBank database and accession numbers are listed in the Supplementary Table S1.
2.4 Phylogenetics and sequence analysis
Seventy-three H1N1pdm clinical samples that yielded sufficient WGS coverage of the IAV genome were included in sequence analysis. In addition to these genomes, we retrieved twenty reference genomes including vaccine strains from the Global Initiative on Sharing All Influenza Data (GISAID, https://www.gisaid.org/) for further analyses (see acknowledgment table in Supplementary Table S2). Nucleotide sequences of the longest open reading frames (ORFs) in each gene segment were used to build phylogenetic trees, including PB2, PB1, PA, HA, NP, NA, M1 and NS1, as well as their alternative splicing (M2 and NS2) and ribosomal frameshift (PA-X) products. Nucleotide sequences of the coding region between the most 5′ start codon and most 3′ stop codon in each segment were separated and concatenated to generate one single sequence for constructing a whole-genome phylogenetic tree. Phylogenetic trees for investigating their genetic or genomic relationships were based on maximum likelihood (ML) analysis, generated with RAxML (version 8.2.12) (Stamatakis 2014) under the GTRGAMMA model with 1,000 bootstrap replicates. Their time scales based on sample collection dates were calibrated using the coalescent model in TreeTime (Sagulenko et al. 2018) after building ML trees. Clades 6B, 6B.1 and 6B.2 were defined by the rules of HA amino acid mutations of global surveillance groups from nextflu (Neher and Bedford 2015). Phylogenetic groups in other gene segments were based on their tree branches to separate annotated clades (e.g. 6B.1 and 6B.2). Inconsistent positioning of strains or clade groups on individual trees was used to identify influenza reassortment. Individual gene segment clades were annotated and visualized using the ggtree R package (Yu et al. 2017).
All H1N1pdm genomes (n = 2,873) isolated from September 2015 to May 2016 were downloaded from GISAID as of January 2020 and were further aligned using MAFFT tool (Katoh and Standley 2013) under the default conditions. Moreover, we retrieved all nucleoprotein (NP) sequences (n = 20,477) of H1N1pdm isolated after the year of 2009 (inclusive) from GISAID as of January 2020 to investigate the population of viruses encoding an upstream translation initiation codon (AUG) in the NP gene segment. 20,470 NP sequences were used for further analyses, after aligning and removing seven sequences with unexpected gaps. Two acknowledgment tables of 2,873 genomes and 20,470 NP sequences were provided (Supplementary Tables S3 and S4, respectively).
2.5 Glycosylation prediction
Potential N-linked glycosylation sites of the 6B.1 and 6B.2 HA and NA proteins were predicted by the NetNGlyc 1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc).
2.6 Statistical analysis of demographic and clinical data
To identify a contribution of virus clades and genetic subgroups to clinical symptoms, we applied chi-square or Fisher's exact tests for categorical variables and the rank-sum test for continuous variables when appropriate. The two-tailed statistical significance was set at P-value <0.05. When clinical or demographic category reached statistical significance in a univariate analysis, confounding effects were then analyzed. In addition to adjusting for possible confounding effects between demographics, disease severity and clinical outcomes, a propensity score-based weighting was applied. Briefly, we applied those potential confounders to generate the propensity (or probability) of being infected with different subgroups of H1N1pdm viruses by using machine learning-based modeling methods (Lee et al. 2010). By using the inverse probability of treatment weighting (IPTW) method, patients with higher propensity will have lower weights in the final multivariable logistic regression model to adjust for confounding factors. The weights create a pseudo-population where the weighted virus subgroups have similar distribution across these confounders. In one simulation model, the propensity score-based weighting methods could adjust for most of the confounding effects (Stuart 2010). Odds ratios of unadjusted and IPTW were calculated using logistic regression analysis. De-identified line-listing data for each case are provided in Supplementary Table S5. All data are available in supplemental figures or in GISAID.
3. Results
3.1 H1N1pdm clade 6B.1 predominantly circulated in Baltimore and clades 6B.1 and 6B.2 co-circulated in Northern Taiwan
From November 2015 to April 2016, 261 and 256 symptomatic patients were enrolled in the study from emergency departments in JHMI (Baltimore) and CGMH (northern Taiwan). IAV was found in 32.6 per cent (85/261) of symptomatic patients from Baltimore and 57.8 per cent (148/256) from northern Taiwan. About half of the IAV-positive samples were further subtyped, and the demographics and comorbidities were found to be similar. In 97.5 per cent (39/40) of samples from Baltimore and 90.1 per cent (64/71) of samples from northern Taiwan H1N1 was the IAV subtype. The epidemic curves of the two surveillance sites were different, with IAV activity starting in January and peaking in mid-March in Baltimore, while IAV activity in northern Taiwan started in November and peaked in late January/early February, with peak activity higher than in Baltimore (Fink et al. 2020).
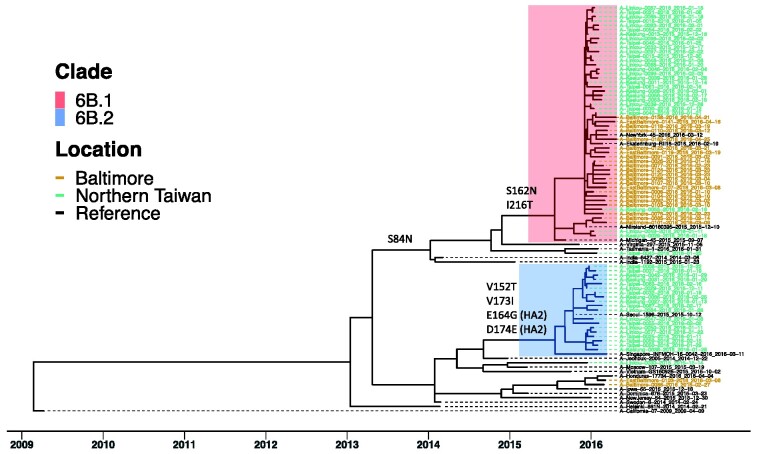
Seventy-three of the 103 H1N1pdm clinical samples yielded sufficient sequence coverage of the IAV genome. Virus samples were initially analyzed based on HA segment sequences (Fig. 1). In Baltimore, 91.7 per cent (22/24) of viruses were clade 6B.1, two were clade 6B and no 6B.2 viruses were identified. The northern Taiwan samples consisted of 57.1 per cent (28/49) clade 6B.1, 38.8 per cent (19/49) clade 6B.2 and 4.1 per cent (2/49) clade 6B viruses. The difference in clade circulation is consistent with global influenza surveillance that H1N1pdm 6B.1 and 6B.2 were dominant in the 2015–16 season and most 6B.2 viruses were found in Asia (Bedford and Neher 2017; World Health Organization 2016).
Figure 1.
H1N1pdm clade 6B.1 and clade 6B.2 circulated in Baltimore and Northern Taiwan in the 2015–16 season. ML tree of HA gene coding sequences from seventy-three H1N1pdm viruses of the surveillance was generated using RAxML under GTRGAMMA model with 1,000 bootstrap replicates. Their time scales based on sample collection dates, noted in each sample, were calibrated using coalescent model in TreeTime after building ML tree. Reference sequences are H1N1pdm vaccine strains, A/California/07/2009 and A/Michigan/45/2015 and three 6B.1, two 6B.2 and thirteen 6B viruses. Tips were colored by location of sample collection. Samples from Baltimore were colored in brown; samples from northern Taiwan were colored in green. Most samples were grouped into two clusters, 6B.1 (in red) and 6B.2 (in blue). Specific amino acid mutations defining major branches of 6B.1 and 6B.2 were indicated on the side of branch.
3.2 Identification of four distinct genetic subgroups of H1N1pdm
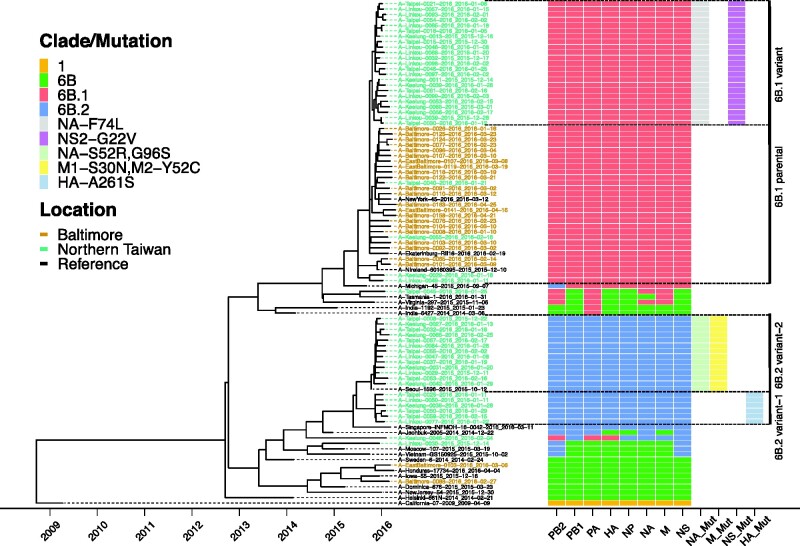
To better understand the genetic diversity in the identified HA clades, we performed a phylogenetic analysis using concatenated gene segments (Fig. 2) and individual ORF trees (Supplementary Fig. S1 and Fig. 2). Branch tips of the ten ORFs from the other seven gene segments were colored by the HA clade of each sample and defining amino acid mutations were noted (Supplementary Fig. S1). For the most part, the sequences of other ORFs clustered together consistent with their HA clade (Fig. 2). Amino acid differences between clades 6B.1 and 6B.2 are listed for HA (Supplementary Table S6), NA (Supplementary Table S7) and the other ORFs (Supplementary Table S8). HA and concatenated trees annotating with confidence intervals of divergence times were provided (Supplementary Figs. S2 and S3), showing the assessment of phylogenetic signals.
Figure 2.
Four distinct genetic subgroups of H1N1pdm were identified in Baltimore and Northern Taiwan in the 2015–16 season. ML trees of concatenated gene segments of 73 H1N1pdm viruses of the surveillance were generated using RAxML under GTRGAMMA model with 1,000 bootstrap replicates. Their time scales based on sample collection dates, noted in each sample, were calibrated using coalescent model in TreeTime after building ML trees. Reference sequences are H1N1pdm vaccine strains, A/California/07/2009 and A/Michigan/45/2015 and three 6B.1, two 6B.2 and thirteen 6B viruses. Tips were colored by location of sample collection. Samples from Baltimore were colored in brown; samples from northern Taiwan were colored in green. A schematic representation of clade/mutation of each gene segment based on their individual gene tree branches to separate annotated clades (1, 6B, 6B.1 or 6B.2) and amino acid mutations (see Supplementary Fig. S1) was annotated and visualized using the ggtree R package. Mutations (Mut) in NA, M, NS and HA segments were identified to be able to divide majority of viruses into four distinct genetic subgroups, 6B.1 parental, 6B.1 variant, 6B.2 variant-1 and 6B.2 variant-2.
Additional amino acid mutations were identified to differentiate viruses into four distinct virus genetic subgroups (Fig. 2). The 6B.1 viruses were separated into two groups, a 6B.1 parental and a 6B.1 variant defined by unique amino acid changes in NA and NS2 (Supplementary Tables S7 and S8). 6B.1 parental viruses were closed to clade 6B.1 viruses from New York, Ekaterinburg and North Ireland. All Baltimore 6B.1 viruses were 6B.1 parental. In northern Taiwan, only four were 6B.1 parental, with the remaining clade 6B.1 viruses (23/27) being 6B.1 variant. The 6B.2 viruses were also separated into two genetic subgroups—variant 1 and variant 2—that are differentiated by mutations in HA, NA, M1 and M2 (Supplementary Tables S6, S7 and S8). 6B.2 variant 1 viruses were very similar to a 6B.2 virus from Singapore; however, the Singapore virus does not contain HA-A261S mutation. 6B.2 variant 2 viruses were clustered with a 6B.2 virus from Seoul, which contains the same mutations in NA, M1 and M2.
3.3 Dynamic virus evolution of gene segments and possible reassortment
All segments in two of the four 6B viruses, A/EastBaltimore/0103/2016 and A/Baltimore/0088/2016, were clade 6B. One 6B virus A/Taipei/0045/2016 contained mutations in PB2, PA, NA and M segments consistent with those found in the clade 6B.1 and the remaining segments were similar but distinct from clade 6B.1. The other 6B virus A/Linkou/0030/2015 contained sequences in the PB2 and NS segments consistent with the clade 6B.2 viruses and the remaining segments were related to 6B.2 but indistinguishable from clade 6B. (Fig. 2 and Supplementary Fig. S1). These data suggest that all segments of ancestor 6B viruses were diverged and evolved toward either the clade 6B.1 or clade 6B.2.
There was also evidence of interclade reassortment in one northern Taiwan 6B.1 sample A/Keelung/0046/2016, as three gene segments of 6B.1 (segments HA, PB2 and PA) and five 6B.2 (segments PB1, NP, NA, M and NS) were detected (Fig. 2). While we cannot rule out co-infection with two H1N1 clade viruses, given the number of reads present for each segment, it seems more likely this individual was infected with an interclade reassortant virus.
3.4 HA and NA N-linked glycosylation
Eight N-linked glycosylation sites on the HA of A/California/07/2009 were previously determined by sequence analysis and verified by mass spectrometry (She et al. 2017). The 6B.1 HA sequence predicts an additional N-linked glycosylation site at 162–164 (N-Q-S) on the Sa antigenic site of the head domain due to an S to N mutation at HA-162 (Supplementary Table S6) (Altman et al. 2019). The NA from the A/California/07/2009 vaccine strain has eight potential N-linked glycosylation sites and six sites were experimentally confirmed (She et al. 2017). Both 6B.1 and 6B.2 viruses gain one additional predicted N-linked glycosylation site at 42–44 (N-Q-S) of the NA stalk and lose one at 386–388 (N-F-S), which lies on the lateral surface of the NA head. In addition, 68.4 per cent (13/19) of 6B.2 viruses with NA-S52R mutation lose one glycosylation site at 50–52 (N-Q-S) on the NA stalk (Supplementary Table S7).
3.5 Antiviral resistance mutations
All sequences contained the amantadine resistance mutation M2-S31N, which is found in the A/California/07/2009 virus. One 6B.2 variant 2 sample from Northern Taiwan, A/Linkou/0029/2015, contained the NA-H275Y, the most common mutation associated with oseltamivir resistance in N1 NA.
3.6 An upstream AUG codon lengthens the NP of H1N1pdm
The NP gene segments of all samples in the study have a new, in frame AUG start codon eighteen nucleotides upstream of the usual start codon of NP proteins of IAV including human seasonal H1N1 and H3N2 (Gorman et al. 1991; Reid et al. 2004), adding six amino acids to the amino terminus of the protein with clade 6B.1 (M-S-D-I-E-A) and 6B.2 viruses (M-S-D-I-E-V) differing at amino acid six. This start codon was shown to be utilized and the resulting NP protein capable of supporting viral polymerase activity (Wanitchang et al. 2011; Wise et al. 2019). The presence of an upstream NP start codon and the amino acid difference at position 6 between 6B.1 and 6B.2 were also present in GISAID. Approximately 70 per cent of GISAID sequences (n = 20,470) of H1N1pdm NP since 2009 were with an Alanine (A) dominating at position 6 and 24 per cent of GISAID sequences (n = 4,924) did not contain genetic data (including A/California/07/2009) in this region. We suspected that the upstream start codon was present in most of the strains when H1N1pdm emerged in 2009.
3.7 Patients infected with clade 6B.2 had increased symptoms of severe influenza
To determine whether viral genotype contributes to symptoms and disease severity, the demographic and clinical data from patients infected with clade 6B.1 or 6B.2 viruses were first compared by univariate analyses (Table 1). Patients infected with 6B.2 were more likely to be older, female, Asian, and unvaccinated. These patients had a higher prevalence of runny nose and diarrhea and had increased symptoms of severe influenza, including increased likelihood of oxygen supplementation, longer in-hospital stay, and pneumonia diagnosis (by radiological findings). Interestingly, patients infected with clade 6B.1 viruses had a lower number of co-morbidities associated with severe influenza, suggesting a healthier population was being infected with those viruses.
Table 1.
Univariate analysis of patients infected with H1N1pdm 6B.1 and 6B.2 viruses of Baltimore and northern Taiwan in the 2015–16 season.
| Characteristic | 6B.1 (n = 49) N (%)/median (IQR) | 6B.2 (n = 19) N (%)/median (IQR) | P value |
|---|---|---|---|
| Demographics | |||
| Site | |||
| JHMI | 22 (44.9) | 0 (0) | <0.0001* |
| CGMH | 27 (55.1) | 19 (100.0) | |
| Age | |||
| In years | 34 (28–44) | 37 (32–52) | 0.20a |
| 18–49 | 40 (81.6) | 12 (63.2) | 0.12 |
| 50–64 | 6 (12.2) | 7 (36.8) | 0.04* |
| 65+ | 3 (6.1) | 0 (0) | 0.55 |
| Gender | |||
| Female | 31 (63.3) | 14 (73.7) | 0.57 |
| Male | 18 (36.7) | 5 (26.3) | |
| Ethnicity | |||
| Non-Hispanic or Latino | 47 (95.9) | 19 (100.0) | 0.99 |
| Hispanic or Latino | 2 (4.1) | 0 (0) | |
| Race | |||
| Black or African American | 16 (32.7) | 0 (0) | <0.01* |
| White | 1 (2.0) | 0 (0) | 0.99 |
| Asian | 27 (55.1) | 19 (100.0) | <0.01* |
| Other | 5 (10.2) | 0 (0) | 0.56 |
| Vaccination and exposure | |||
| Human exposure | 11 (22.4) | 2 (10.5) | 0.31 |
| Travel in last 30 days | 9 (18.4) | 6 (31.6) | 0.33 |
| Influenza vaccine | 8 (16.3) | 0 (0) | 0.09 |
| Co-morbidities and high risk | |||
| No co-morbidities | 26 (53.1) | 7 (36.8) | <0.01* |
| One co-morbidity | 15 (30.6) | 8 (42.1) | 0.4 |
| Multiple co-morbidities | 8 (16.3) | 4 (21.1) | 0.73 |
| Immunosuppressive medication | 1 (2.0) | 0 (0) | 0.99 |
| Pregnant | 1 (2.0) | 0 (0) | 0.99 |
| Disease severity | |||
| NEWS score | 3 (0–6) | 3 (0–6) | 0.81a |
| Oxygen supplementation | 2 (4.1) | 3 (15.8) | 0.13 |
| BiPAP/CPAP | 1 (2.0) | 0 (0) | 0.99 |
| Intubated | 0 (0) | 0 (0) | 0.99 |
| Pneumonia | 7 (14.3) | 7 (36.8) | 0.05* |
| Admitted or observation | 6 (12.2) | 2 (10.5) | 0.99 |
| LOS | 7.0 (1–12) | 10.0 (5–15) | 0.14a |
| ICU | 0 (0) | 0 (0) | 0.99 |
| Death | 0 (0) | 0 (0) | 0.99 |
| Follow-up due to sequala | 3 (6.1) | 1 (5.3) | 0.99 |
| Viral co-infection | 0 (0) | 0 (0) | 0.99 |
| Symptoms | |||
| Fever | 47 (95.9) | 19 (100.0) | 0.99 |
| Cough | 48 (98.0) | 19 (100.0) | 0.99 |
| Sputum | 32 (65.3) | 14 (73.7) | 0.77 |
| Increased sputum | 17 (34.7) | 6 (31.6) | 0.78 |
| Shortness of breath | 35 (71.4) | 10 (52.6) | 0.16 |
| Wheezing | 16 (32.7) | 10 (52.6) | 0.16 |
| Headache | 40 (81.6) | 18 (94.7) | 0.26 |
| Sore throat | 39 (79.6) | 15 (78.9) | 0.99 |
| Body aches | 43 (87.8) | 19 (100.0) | 0.17 |
| Runny nose | 37 (75.5) | 19 (100.0) | 0.02* |
| Sinus pain | 13 (26.5) | 1 (5.3) | 0.091 |
| Fatigue | 43 (87.8) | 18 (94.7) | 0.66 |
| Chest pain | 30 (61.2) | 8 (42.1) | 0.18 |
| Chills | 38 (77.6) | 16 (84.2) | 0.74 |
| Loss of appetite | 36 (73.5) | 16 (84.2) | 0.53 |
| Nausea/vomiting | 28 (57.1) | 12 (63.2) | 0.79 |
| Diarrhea | 10 (20.4) | 9 (47.4) | 0.04* |
| Conjunctivitis | 11 (22.4) | 2 (10.5) | 0.33 |
Statistical analysis was done using R software.
Categorical variables were compared using chi-square or Fisher's exact tests.
Continuous variables were compared with rank-sum test.
Statistical significance was set at P < 0.05 and was represented in Bold and Italics.
We further utilized a logistic regression analysis to estimate the propensity of being infected by 6B.2, using potential confounders such as age, gender, comorbidities, obesity, vaccine status, human exposure and travel history (area under the ROC curve: 0.798). After IPTW to adjust for the possible confounding effects, infection with clade 6B.2 viruses was found to be significantly associated with pneumonia (odds ratio: 3.261, 95% confidence interval (CI): 1.696–4.826, P = 0.008) (Table 2). In order to rule out the possibility of institutional bias because 6B.2 infection was not detected in Baltimore, we excluded Baltimore patients and only compared 6B.1 and 6B.2 of Northern Taiwan patients. The pneumonia odds ratio of IPTW adjusted with the same confounders of 6B.2 over 6B.1 patients in northern Taiwan was still borderline significant (odds ratio: 2.569, 95% CI: 1.026–7.12, P = 0.054) (Supplementary Table S9).
Table 2.
Logistic regression analysis of pneumonia in patients infected with H1N1pdm 6B.1 and 6B.2 viruses of Baltimore and Northern Taiwan in the 2015–16 season.
| Unadjusted |
IPTWa |
||||||
|---|---|---|---|---|---|---|---|
| Clade | Pneumonia | Odds ratio | 95% CI | P value | Odds ratio | 95% CI | P value |
| 6B.1 | 14.3% (7/49) | 1 | – | – | 1 | – | – |
| 6B.2 | 36.8% (7/19) | 3.500 | 1.629–5.372 | 0.046* | 3.261 | 1.696–4.826 | 0.008* |
IPTW results were done by propensity score weighting with recursive partitioning algorithm (area under the ROC curve: 0.798) for age, gender, comorbidities, obesity, vaccine status, human exposure and travel history, then followed by logistic regression analysis.
P < 0.05, logistic regression analysis.
The distribution of cases across the four genetic subgroups was then determined (Tables 3 and 4). Patients infected with the 6B.1 parental virus had increased reporting of wheezing, sinus pain and nausea/vomiting, along with the expected difference in race and ethnicity stemming from the identification of the 6B.1 variant only in northern Taiwan (Table 3). When patients infected with the 6B.2 variant 1 and variant 2 viruses were compared there were no obvious difference in symptoms (Table 4); however, the small sample sizes in the 6B.2 subgroups precluded a definitive characterization of demographic and clinical data between these genotypes. We did not observe difference in pneumonia of patients infected with 6B.1 variant, which only occurred in Northern Taiwan compared with patients with 6B.1 parental infection (mostly Baltimore) (Table 3), suggesting there is not a systematic difference of pneumonia diagnosis between Baltimore and Northern Taiwan. Overall, our data suggested clade 6B.2 circulation in Northern Taiwan likely played a role in differentiating the influenza epidemics between Baltimore and Northern Taiwan in the season.
Table 3.
Univariate analysis of patients infected with H1N1pdm 6B.1 parental and 6B.1 variant viruses of Baltimore and Northern Taiwan in the 2015–16 season.
| Characteristic | 6B.1 Parental (n = 26) N (%)/median (IQR) | 6B.1 Variant (n = 23) N (%)/median (IQR) | P value |
|---|---|---|---|
| Demographics | |||
| Site | |||
| JHMI | 22 (84.6) | 0 (0) | <0.0001* |
| CGMH | 4 (15.4) | 23 (100.0) | |
| Age | |||
| In years | 33 (29–44) | 35 (29–44) | 0.75a |
| 18–49 | 20 (76.9) | 20 (87.0) | 0.47 |
| 50–64 | 5 (19.2) | 1 (4.3) | 0.19 |
| 65+ | 1 (3.8) | 2 (8.7) | 0.59 |
| Gender | |||
| Female | 14 (53.8) | 17 (73.9) | 0.24 |
| Male | 12 (46.2) | 6 (26.1) | |
| Ethnicity | |||
| Non-Hispanic or Latino | 24 (92.3) | 23 (100.0) | 0.49 |
| Hispanic or Latino | 2 (7.7) | 0 (0) | |
| Race | |||
| Black or African American | 16 (61.5) | 0 (0) | <0.0001* |
| White | 1 (3.8) | 0 (0) | 0.99 |
| Asian | 4 (15.4) | 23 (100.0) | <0.0001* |
| Other | 5 (19.3) | 0 (0) | 0.08 |
| Vaccination and exposure | |||
| Human exposure | 4 (15.4) | 7 (30.4) | 0.36 |
| Travel in last 30 days | 3 (11.5) | 6 (26.1) | 0.35 |
| Influenza vaccine | 6 (23.1) | 2 (8.7) | 0.33 |
| Co-morbidities and high risk | |||
| No co-morbidities | 11 (42.3) | 15 (65.2) | 0.15 |
| One co-morbidity | 8 (30.8) | 7 (30.4) | 0.99 |
| Multiple co-morbidities | 7 (26.9) | 1 (4.3) | 0.05 |
| Immunosuppressive medication | 1 (3.8) | 0 (0) | 0.99 |
| Pregnant | 1 (3.8) | 0 (0) | 0.99 |
| Disease severity | |||
| NEWS score | 3 (0–6) | 3 (0–6) | 0.57a |
| Oxygen supplementation | 2 (7.7) | 0 (0) | 0.49 |
| BiPAP/CPAP | 1 (3.8) | 0 (0) | 0.49 |
| Intubated | 0 (0) | 0 (0) | 0.99 |
| Pneumonia | 3 (11.5) | 4 (17.4) | 0.69 |
| Admitted or observation | 4 (15.4) | 2 (8.7) | 0.67 |
| LOS | 6.5 (1–12) | 6.0 (3–9) | 0.67a |
| ICU | 0 (0) | 0 (0) | 0.99 |
| Death | 0 (0) | 0 (0) | 0.99 |
| Follow-up due to sequala | 2 (7.7) | 1 (4.3) | 0.99 |
| Viral co-infection | 0 (0) | 0 (0) | 0.99 |
| Symptoms | |||
| Fever | 24 (92.3) | 23 (100.0) | 0.49 |
| Cough | 25 (96.2) | 23 (100.0) | 0.99 |
| Sputum | 15 (57.7) | 17 (73.9) | 0.37 |
| Increased sputum | 10 (38.5) | 7 (30.4) | 0.77 |
| Shortness of breath | 20 (76.9) | 15 (65.2) | 0.53 |
| Wheezing | 12 (46.2) | 4 (17.4) | 0.04* |
| Headache | 23 (88.5) | 17 (73.9) | 0.27 |
| Sore throat | 19 (73.1) | 20 (87.0) | 0.3 |
| Body aches | 24 (92.3) | 19 (82.6) | 0.4 |
| Runny nose | 17 (65.4) | 20 (87.0) | 0.1 |
| Sinus pain | 11 (42.3) | 2 (8.7) | 0.01* |
| Fatigue | 22 (84.6) | 21 (91.3) | 0.67 |
| Chest pain | 19 (73.1) | 11 (47.8) | 0.08 |
| Chills | 21 (80.8) | 17 (73.9) | 0.73 |
| Loss of appetite | 20 (76.9) | 16 (69.6) | 0.75 |
| Nausea/vomiting | 19 (73.1) | 9 (39.1) | 0.02* |
| Diarrhea | 7 (26.9) | 3 (13.0) | 0.3 |
| Conjunctivitis | 8 (30.8) | 3 (13.0) | 0.18 |
Statistical analysis was done using R software.
Categorical variables were compared using chi-square or Fisher's exact tests.
Continuous variables were compared with rank-sum test.
Statistical significance was set at P < 0.05 and was represented in Bold and Italics.
Table 4.
Univariate analysis of patients infected with H1N1pdm 6B.2 variant 1 and 6B.2 variant 2 viruses of Baltimore and Northern Taiwan in the 2015–16 season.
| Characteristic | 6B.2 Variant-1 (n = 6) N (%)/median (IQR) | 6B.2 Variant-2 (n = 13) N (%)/median (IQR) | P value |
|---|---|---|---|
| Demographics | |||
| Site | |||
| JHMI | 0 (0) | 0 (0) | 0.99 |
| CGMH | 6 (100.0) | 13 (100.0) | |
| Age | |||
| In years | 34 (29–48) | 44 (34–54) | 0.20a |
| 18–49 | 4 (66.7) | 8 (61.5) | 0.99 |
| 50–64 | 2 (33.3) | 5 (38.5) | 0.99 |
| 65+ | 0 (0) | 0 (0) | 0.99 |
| Gender | |||
| Female | 3 (50.0) | 11 (84.6) | 0.26 |
| Male | 3 (50.0) | 2 (15.4) | |
| Ethnicity | |||
| Non-Hispanic or Latino | 6 (100.0) | 13 (100.0) | 0.99 |
| Hispanic or Latino | 0 (0) | 0 (0) | |
| Race | |||
| Black or African American | 0 (0) | 0 (0) | 0.99 |
| White | 0 (0) | 0 (0) | 0.99 |
| Asian | 6 (100.0) | 13 (100.0) | 0.99 |
| Other | 0 (0) | 0 (0) | 0.99 |
| Vaccination and exposure | |||
| Human exposure | 0 (0) | 2 (15.4) | 0.83 |
| Travel in last 30 days | 3 (50.0) | 3 (23.1) | 0.32 |
| Influenza vaccine | 0 (0) | 0 (0) | 0.99 |
| Co-morbidities and high risk | |||
| No co-morbidities | 3 (50.0) | 4 (30.8) | 0.77 |
| One co-morbidity | 2 (33.3) | 6 (46.2) | 0.99 |
| Multiple co-morbidities | 1 (16.7) | 3 (23.1) | 0.99 |
| Immunosuppressive medication | 0 (0) | 0 (0) | 0.99 |
| Pregnant | 0 (0) | 0 (0) | 0.99 |
| Disease severity | |||
| NEWS score | 4 (2–6) | 3 (0–5) | 0.54a |
| Oxygen supplementation | 0 (0) | 3 (23.1) | 0.52 |
| BiPAP/CPAP | 0 (0) | 0 (0) | 0.99 |
| Intubated | 0 (0) | 0 (0) | 0.99 |
| Pneumonia | 2 (33.3) | 5 (38.5) | 0.99 |
| Admitted or observation | 0 (0) | 2 (15.4) | 0.99 |
| LOS | 0 (0 –0) | 10.0 (5 –15) | <0.01* |
| ICU | 0 (0) | 0 (0) | 0.99 |
| Death | 0 (0) | 0 (0) | 0.99 |
| Follow-up due to sequala | 0 (0) | 1 (7.7) | 0.99 |
| Viral co-infection | 0 (0) | 0 (0) | 0.99 |
| Symptoms | |||
| Fever | 6 (100.0) | 13 (100.0) | 0.99 |
| Cough | 6 (100.0) | 13 (100.0) | 0.99 |
| Sputum | 5 (83.3) | 9 (69.2) | 0.99 |
| Increased sputum | 3 (50.0) | 3 (23.1) | 0.32 |
| Shortness of breath | 2 (33.3) | 8 (61.5) | 0.35 |
| Wheezing | 2 (33.3) | 8 (61.5) | 0.35 |
| Headache | 6 (100.0) | 12 (92.3) | 0.99 |
| Sore throat | 6 (100.0) | 9 (69.2) | 0.26 |
| Body aches | 6 (100.0) | 13 (100.0) | 0.99 |
| Runny nose | 6 (100.0) | 13 (100.0) | 0.99 |
| Sinus pain | 0 (0) | 1 (7.7) | 0.99 |
| Fatigue | 6 (100.0) | 12 (92.3) | 0.99 |
| Chest pain | 2 (33.3) | 6 (46.2) | 0.99 |
| Chills | 5 (83.3) | 11 (84.6) | 0.99 |
| Loss of appetite | 6 (100.0) | 10 (76.9) | 0.52 |
| Nausea/vomiting | 3 (50.0) | 9 (69.2) | 0.62 |
| Diarrhea | 4 (66.7) | 5 (38.5) | 0.35 |
| Conjunctivitis | 0 (0) | 2 (15.4) | 0.99 |
Statistical analysis was done using R software.
Categorical variables were compared using Chi-square or Fisher's exact tests.
Continuous variables were compared with rank-sum test.
Statistical significance was set at p < 0.05 and was represented in Bold and Italics.
4. Discussion
Four distinct genetic subgroups of H1N1pdm viruses were circulating in Baltimore and northern Taiwan during the 2015–16 flu season. While the 6B.1 clade was circulating at both sites, some 6B.1 viruses in Northern Taiwan had additional mutations that defined a genetic subgroup we called 6B.1 variant. The two 6B.1 subgroups of viruses did not differ drastically with respect to disease severity. The 6B.2 clade viruses were detected in Northern Taiwan but not in Baltimore and consisted of two distinct genetic subgroups. Patients infected with 6B.2 virus showed more and stronger disease symptoms by both uni- and multivariate analysis. This difference in H1N1pdm clade circulation may have contributed to the more severe influenza season experienced in Northern Taiwan compared to Baltimore in 2015–16. After adjusting and weighting for age, gender, comorbidities, obesity, vaccine status, human exposure and travel history, people infected with 6B.2 had a slightly lower but still statistically significant odds developing pneumonia (Table 2). The data indicate that infection with clade 6B.2 viruses is associated with a higher disease severity at our surveillance sites independent of patient demographics and comorbidities but different race populations were difficult to adjust for. The presence of the 6B.2 clade in addition to low preexisting immunity to circulating H1N1 in Northern Taiwan (Fink et al. 2020) may be the most likely reasons explaining the different numbers and severity of influenza cases between Baltimore and Northern Taiwan. The only report for symptoms of 6B.1 compared with 6B.2 infection was seen in Israel, where infection with 6B.2 virus was associated with more vomiting and nausea compared to 6B.1-infected patients (Friedman et al. 2017). In our surveillance, we did see a higher percentage of 6B.2 than 6B.1 patients with vomiting and nausea, but this did not reach statistical significance (Table 1).
The H1N1pdm 6B.1 and 6B.2 viruses dominated the 2015–16 season in the Northern Hemisphere. However, numbers of cases and disease severity differed between geographic regions. The 2015–16 H1N1pdm season in the United States and Canada were moderate compared with the prior season (Chambers et al. 2016; Davlin et al. 2016). On the other hand, influenza surveillance in Russia, UK, eastern Europe, the Middle East and Asia (including Taiwan) had increasing numbers of severe cases (Komissarov et al. 2016; National Institute of Infectious Diseases Japan 2016; Pebody et al. 2016; Newitt et al. 2018; Public Health England (PHE) 2016; Taiwan CDC 2016; Tjon-Kon-Fat et al. 2016). Reports also showed H1N1pdm impacted the southern hemisphere in the 2016 season, in particular, Brazil and Reunion Island (Filleul et al. 2016; Santos et al. 2017; Cardoso et al. 2019).
Serological surveillance indicated that the circulating H1N1pdm strains in 2015–16 were antigenically similar to the vaccine A/California/07/2009 in the United States, United Kingdom and many other countries (Broberg et al. 2016; Chambers et al. 2016; Davlin et al. 2016; Komissarov et al. 2016; Pebody et al. 2016; Korsun et al. 2017). This suggests that the HA amino acid differences between the circulating 6B.1 or 6B.2 viruses and the vaccine strain were likely not able to explain differential epidemics in different regions of the year even though vaccination coverage might differ. Whether mutations in the other seven viral genes contributed to the variable disease severity reported in 2015–16 has not been studied carefully. Our whole-genome analysis data showed there were sixteen amino acid differences in the other 8 viral ORFs (no differences in PB1 or M2) between clade 6B.1 and 6B.2 viruses (Supplementary Tables S7 and S8). Some differences in NA and M1 occurred at sites previously associated with altered virus transmission (Bialas et al. 2012; Lv et al. 2015), but our initial experiments on human nasal epithelial cell cultures did not detect any replication differences between clade 6B.1 and 6B.2 viruses (Fink et al. 2020).
We identified one 6B.2 patient in northern Taiwan had NA-H275Y mutation, which is associated with NA drug resistance. In another study of 2015–16 H1N1pdm in Taiwan, one clinical isolate with NA-H275Y was resistant to oseltamivir (Gong et al. 2019). The data suggest sporadic identification of oseltamivir resistant H1N1pdm viruses in 2015–16 in Taiwan.
Severe and fatal outbreaks occurred in early 2015 in Nepal and India (Nakamura et al. 2017). A study in Taiwan showed patients with laboratory-confirmed H1N1pdm in 2013–14 and 2015–16 6B, 6B.1 and 6B.2 seasons had higher risk for influenza-related complications compared with patients from seasons where 6B, 6B.1 and 6B.2 viruses did not dominate (Hsieh et al. 2018). Both suggested 6B viruses, ancestors of 6B.1 and 6B.2, may have caused severe outcomes.
The clade 6B.1 parental genetic subgroup continued to circulate after 2015–16, and evolved into several 6B.1A subclades through August 2020. It is not clear what drove the extinction of 6B.2 but it is possible that clade 6B.1 viruses were better adapted to infect and spread in humans. It has been suggested that glycosylation is important for IAV adaptation in humans (Tate et al. 2014) and gaining a potential glycosylation site on the HA head at residue 162 may have given clade 6B.1 viruses an evolutionary advantage. Clade 6B.1 viruses also encode an NS1-E125D mutation which increases NS1 interactions with cellular cleavage and polyadenylation factor 30 (CPSF30) (Hale et al. 2010). The NS1-E125D mutation of 6B.1 viruses has been suggested to be an important marker for virus adaption to humans (Komissarov et al. 2016).
Our study is limited in several ways. The different race distributions between our surveillance sites make it difficult to adjust for in this and any study conducted across different geographic regions. The identification of 4 genetic subgroups reduced the power of our study to detect demographic and clinical differences in our populations. Increasing surveillance and whole-genome sequencing efforts could dramatically improve the ability to identify novel virus variants that are causing altered disease in humans. Given these limitations, it is still clear that there is significant H1N1 genetic diversity across the two surveillance sites and this genetic diversity contributes to the differing number of influenza cases and disease severity. Influenza is a global disease, but surveillance at the local level is needed to fully understand virus circulation and disease potential.
Supplementary Material
Acknowledgements
The authors thank the patients who enrolled and participated in the JH-CEIRS surveillance study. We are grateful for the efforts of the clinical coordination team at JHMI and CGMH who collected samples. We thank members of the Pekosz lab for feedback on this work.
Supplementary data
Supplementary data are available at Virus Evolution online.
Author contributions
H.L., R.R., K.F.C. and A.P. conceived of the experimental questions; Y.N.G. performed phylogenetic tree analyses; K.S.S. and K.F.C. ran demographic and clinical analyses; T.M., J.E. and P.T. sequenced samples and cleaned the raw data; Z.Y.L., M.L. and L.S. oversaw enrollment and collection of samples from patients; H.L., Y.N.G., K.S.S. and K.F.C. analyzed data; H.L. and A.P. wrote the manuscript; all authors edited and reviewed the manuscript prior to submission.
Funding
This work was supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases Center of Excellence in Influenza Research and Surveillance contract HHS N2772201400007C (R.R., K.F.C., and A.P.), the Richard Eliasberg Family Foundation, the Research Center for Emerging Viral Infections from the Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan, the Ministry of Science and Technology (MOST), 109-2634-F-182-001 (Y.N.G.) and 109-2327-B-182-002 (K.F.C.), and Chang Gung Memorial Hospital in Taiwan CMRPG2H0323, CMRPG2H0312, CMRPG2K0211, and CMRPG2K0241 (K.F.C.)..
Conflict of interest: None declared.
References
- Altman M. O. et al. (2019) ‘Human Influenza a Virus Hemagglutinin Glycan Evolution Follows a Temporal Pattern to a Glycan Limit’, mBio, 10: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedford T., Neher R. (2017), 'Seasonal Influenza Circulation Patterns and Projections for 2017-2018’, <https://nextflu.org/reports/feb-2017/> accessed 27 Sep 2020.
- Bialas K. M., Desmet E. A., Takimoto T. (2012) ‘Specific Residues in the 2009 H1N1 Swine-Origin Influenza Matrix Protein Influence Virion Morphology and Efficiency of Viral Spread in Vitro’, PLoS One, 7: e50595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier N. M., Palese P. (2008) 'The Biology of Influenza Viruses’, Vaccine, 26 Suppl 4: D49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broberg E. et al. (2016) 'Predominance of Influenza A(H1N1)pdm09 Virus Genetic Subclade 6B.1 and Influenza B/Victoria Lineage Viruses at the Start of the 2015/16 Influenza Season in Europe’, Eurosurveillance, 21(13). doi: 10.2807/1560-7917.ES.2016.21.13.30184. [DOI] [PubMed] [Google Scholar]
- Cardoso A. M. et al. (2019) ‘Investigation of an Outbreak of Acute Respiratory Disease in an Indigenous Village in Brazil: Contribution of Influenza A(H1N1)pdm09 and Human Respiratory Syncytial Viruses’, PLoS One, 14: e0218925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers C. et al. (2016) ‘Interim Estimates of 2015/16 Vaccine Effectiveness against Influenza A(H1N1)pdm09, Canada, February 2016’, Eurosurveillance, 21: 30168. [DOI] [PubMed] [Google Scholar]
- Davlin S. L. et al. (2016) ‘ 'Influenza Activity - United States, 2015-16 Season and Composition of the 2016-17 Influenza Vaccine’, Morbidity and Mortality Weekly Report, 65: 567–75. [DOI] [PubMed] [Google Scholar]
- Filleul L. et al. (2016) ‘A Major Impact of the Influenza Seasonal Epidemic on Intensive Care Units, Reunion, April to August ’, Eurosurveillance, 21: 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink A. L. et al. (2020), ‘Regional Differences in Vaccine Uptake and Serological Responses to Vaccine and Circulating Strains of H1N1 Viruses among Patients with Confirmed Influenza’, medRxiv, 2020.10.03.20203042.
- Friedman N. et al. (2017) ‘A(H1N1)pdm09 Influenza Infection: Vaccine Inefficiency’, Oncotarget, 8: 32856–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garretson T. A. et al. (2018) ‘ 'Identification of Human Vaccinees That Possess Antibodies Targeting the Egg-Adapted Hemagglutinin Receptor Binding Site of an H1N1 Influenza Vaccine Strain’, Vaccine, 36: 4095–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y. N. et al. (2018) 'Centennial Review of Influenza in Taiwan’, Biomed J, 41: 234–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y. N. et al. (2019) ‘Population Dynamics at Neuraminidase Position 151 of Influenza A (H1N1)pdm09 Virus in Clinical Specimens’, Journal of General Virology, 100: 752–9. [DOI] [PubMed] [Google Scholar]
- Gorman O. T. et al. (1991) ‘Evolution of Influenza a Virus Nucleoprotein Genes: Implications for the Origins of H1N1 Human and Classical Swine Viruses’, Journal of Virology, 65: 3704–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale B. G. et al. (2010) ‘Inefficient Control of Host Gene Expression by the 2009 Pandemic H1N1 Influenza a Virus NS1 Protein’, Journal of Virology, 84: 6909–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris P. A. et al. (2009) ‘Research Electronic Data Capture (REDCap)–a Metadata-Driven Methodology and Workflow Process for Providing Translational Research Informatics Support’, Journal of Biomedical Informatics, 42: 377–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris P. A. et al. (2019) ‘The REDCap Consortium: Building an International Community of Software Platform Partners’, Journal of Biomedical Informatics, 95: 103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh Y. C. et al. (2018) ‘Clinical Characteristics of Patients with Laboratory-Confirmed Influenza A(H1N1)pdm09 during the 2013/2014 and 2015/2016 Clade 6B/6B.1/6B.2-Predominant Outbreaks’, Scientific Reports, 8: 15636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013) ‘MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability’, Molecular Biology and Evolution, 30: 772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarov A. et al. (2016) ‘Rapid Spread of Influenza A(H1N1)pdm09 Viruses with a New Set of Specific Mutations in the Internal Genes in the Beginning of 2015/2016 Epidemic Season in Moscow and Saint Petersburg (Russian Federation)’, Influenza and Other Respiratory Viruses, 10: 247–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsun N. et al. (2017) ‘Antigenic and Genetic Characterization of Influenza Viruses Circulating in Bulgaria during the 2015/2016 Season’, Infection, Genetics and Evolution, 49: 241–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S. L. (2012) ‘Fast Gapped-Read Alignment with Bowtie 2’, Nature Methods, 9: 357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B. K., Lessler J., Stuart E. A. (2010) ‘Improving Propensity Score Weighting Using Machine Learning’, Statistics in Medicine, 29: 337–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. (2009) ‘The Sequence Alignment/Map Format and SAMtools’, Bioinformatics, 25: 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J. et al. (2015) ‘Amino Acid Substitutions in the Neuraminidase Protein of an H9N2 Avian Influenza Virus Affect Its Airborne Transmission in Chickens’, Veterinary Research, 46: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K. et al. (2017) ‘Characterization of Influenza A(H1N1)pdm09 Viruses Isolated from Nepalese and Indian Outbreak Patients in Early 2015’, Influenza and Other Respiratory Viruses, 11: 399–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute of Infectious Diseases Japan (2016) ‘Influenza 2015/16 Season, Japan’, Infectious Agents Surveillance Report, 37: 211–3. [Google Scholar]
- Neher R. A., Bedford T. (2015) ‘Nextflu: Real-Time Tracking of Seasonal Influenza Virus Evolution in Humans’, Bioinformatics, 31: 3546–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newitt S. et al. (2018) ‘Rapid Risk Assessment during the Early Weeks of the 2015-2016 Influenza Season in Ukraine’, Influenza and Other Respiratory Viruses, 12: 241–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pebody R. et al. (2016) ‘Effectiveness of Seasonal Influenza Vaccine in Preventing Laboratory-Confirmed Influenza in Primary Care in the United Kingdom: 2015/16 Mid-Season Results’, Eurosurveillance, 21. [DOI] [PubMed] [Google Scholar]
- Public Health England (PHE) (2016) 'Summary of UK Surveillance of Influenza and Other Seasonal Respiratory Illnesses 26 May 2016–Week 21 Report (Up To Week 20 Data)’, PHE weekly National Influenza report. <https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/681012/Weekly_national_influenza_report_week_21_2016.pdf> accessed 27 Sep 2020.
- Reid A. H. et al. (2004) ‘Novel Origin of the 1918 Pandemic Influenza Virus Nucleoprotein Gene’, Journal of Virology, 78: 12462–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagulenko P., Puller V., Neher R. A. (2018) ‘TreeTime: Maximum-Likelihood Phylodynamic Analysis’, Virus Evolution, 4: vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos K. C. et al. (2017) ‘Molecular Epidemiology of Influenza A(H1N1)PDM09 Hemagglutinin Gene Circulating in Sao Paulo State, Brazil: 2016 Anticipated Influenza Season’, The Revista do Instituto de Medicina Tropical de São Paulo, 59: e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Y. M. et al. (2017) ‘Topological N-Glycosylation and Site-Specific N-Glycan Sulfation of Influenza Proteins in the Highly Expressed H1N1 Candidate Vaccines’, Science Report, 7: 10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014) ‘RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies’, Bioinformatics, 30: 1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart E. A. (2010) ‘Matching Methods for Causal Inference: A Review and a Look Forward’, Statistics in Medicine, 25: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiwan CDC (2016) ‘Taiwan Influenza Express 2015-16 Flu Season 2016 Week 20’, <https://www.cdc.gov.tw/En/File/Get/VCNkIvtTOiuP1vo1gJG89A> accessed 27 Sep 2020.
- Tate M. D. et al. (2014) ‘Playing Hide and Seek: How Glycosylation of the Influenza Virus Hemagglutinin Can Modulate the Immune Response to Infection’, Viruses, 6: 1294–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjon-Kon-Fat R. et al. (2016) ‘The Potential Risks and Impact of the Start of the 2015-2016 Influenza Season in the WHO European Region: A Rapid Risk Assessment’, Influenza and Other Respiratory Viruses, 10: 236–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanitchang A. et al. (2011) ‘Atypical Characteristics of Nucleoprotein of Pandemic Influenza Virus H1N1 and Their Roles in Reassortment Restriction’, Archives Virology, 156: 1031–40. [DOI] [PubMed] [Google Scholar]
- Wise H. M. et al. (2019) ‘An Alternative AUG Codon in Segment 5 of the 2009 Pandemic Influenza A Virus is a Swine-Derived Virulence Motif’, bioRxiv, 738427.
- World Health Organization (2016) ‘Recommended Composition of Influenza Virus Vaccines for Use in the 2016-2017 Northern Hemisphere Influenza Season’, Wkly Epidemiol Rec, 91: 121–32. [PubMed] [Google Scholar]
- Yu G. et al. (2017) ‘Ggtree: An r Package for Visualization and Annotation of Phylogenetic Trees with Their Covariates and Other Associated Data’, Methods in Ecology and Evolution, 8: 28–36. [Google Scholar]
- Zhou B. et al. (2009) ‘Single-Reaction Genomic Amplification Accelerates Sequencing and Vaccine Production for Classical and Swine Origin Human Influenza a Viruses’, Journal of Virology, 83: 10309–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.