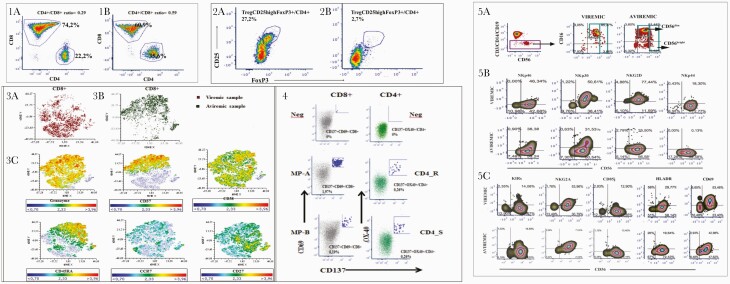
Figure 2.
Panel 1, Comparison of CD4+/CD8+ ratio in CD3+ T cells derived from viremic (panel 1A) and aviremic (panel 1B) samples. Panel 2, Comparison of CD4+FoxP3+CD25+ Treg cells in CD4+ T cells derived from viremic (panel 2A) and aviremic (panel 2B) samples. Panel 3, t-SNE analysis to the multiparametric analyses performed on CD8+ T cells in both samples. Panel 3A, t-SNE color plot of CD8+ lymphocytes in viremic samples. Panel 3B, t-SNE plots of concatenated CD8+ lymphocytes in aviremic samples. Panel 3C, description of phenotypic distributions to maturation and senescence markers, such as CD45RA, CCR7, CD27, CD38, CD57, and granzyme expression. Panel 4, FACS plot analysis of SARS-CoV-2-specific CD8+ and CD4+ T-cell responses of aviremic sample. To measure SARS-CoV-2-specific CD4+ and CD8+ T cells, peripheral blood mononuclear cells were stimulated with a spike MP (MP_S) and the class II MP representing all the proteomes without spike (“nonspike,” MP CD4_R) and HLA A and HLA B peptide megapools (MP A, MP B). SARS-CoV-2-specific CD4+ T-cell responses have been expressed as frequency of CD137+OX40+ cells on total CD4+ T-cell population, whereas SARS-CoV-2-specific CD8+ T-cell responses have been expressed as frequency of CD137+CD69+ cells on total CD8+ T-cell population. Panel 5, Flow cytometric analysis of NK cells at viremic or aviremic time points. Panel 5A, Gating strategy to identify peripheral blood NK cells. Panel 5B, Flow cytometric analysis of the expression of activating (NKp46, NKp30, NKp44, NKG2D) NK cell receptors. Panel 5C, Flow cytometric analysis of inhibitory NK cell receptors (KIRs, NKG2A, CD85j) and activation markers (HLADR, CD69) expressed on the surface of NK cells circulating in peripheral blood in the presence or absence of viremia. To identify SARS-CoV-2-specific CD8+ T cells, 2 class I peptide MPs have been used, based on epitope predictions for the 12 most common HLA A and B alleles, which collectively encompass 628 predicted HLA class I CD8+ T-cell epitopes from the entire SARS-CoV-2 proteome (CD8 MP-A and MP-B).