Fig. 4.
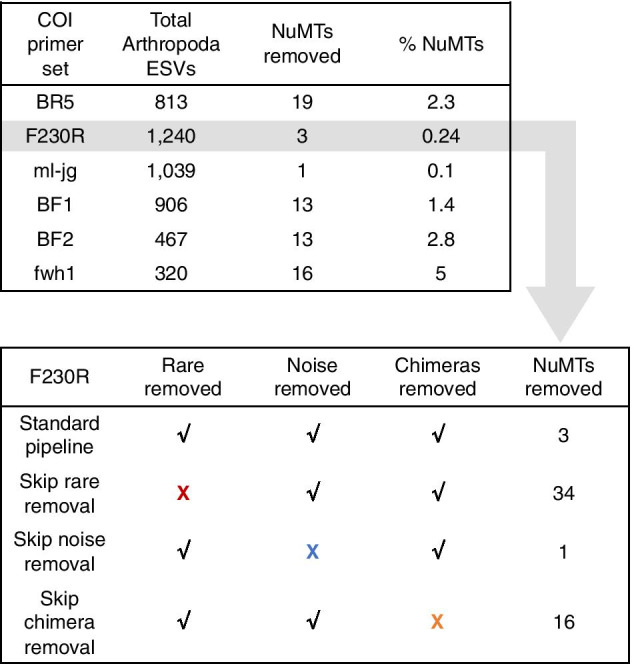
Removing rare sequences also removes apparent pseudogenes. The number of removed putative nuMTs was calculated for each of the 5 amplicons from a freshwater COI metabarcode dataset. Note, that we only compared results across Arthropoda ESVs. Using the SCVUC v4.3.0 bioinformatic pipeline, the F230R amplicon recovered the greatest ESV richness (top box) so we used this as a test case for further tests (bottom box). To determine whether current bioinformatic processing steps already help to remove apparent pseudogenes, we dropped one step at a time: removal of rare sequences, removal of noisy sequences, and removal of chimeric sequences
