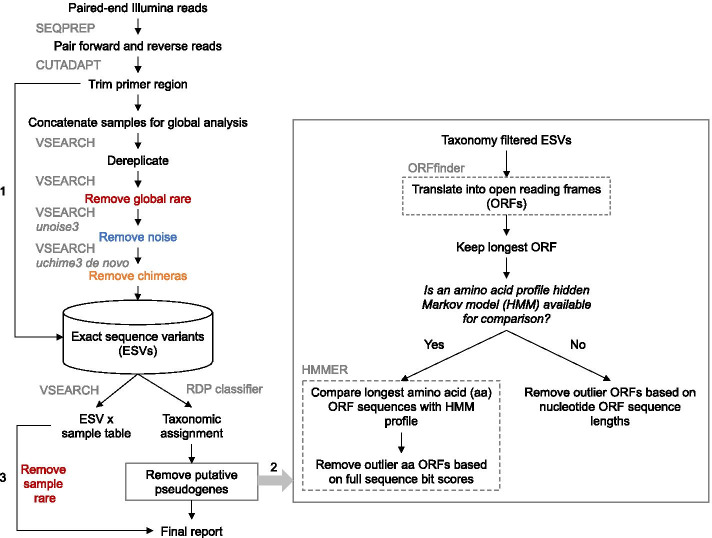
Fig. 5.
Overview of the metabarcoding bioinformatic pipeline that removes apparent pseudogenes. The SCVUC pipelines begins with Illumina paired-end reads. Arrow 1 indicates where primer trimmed reads are mapped to denoised exact sequence variants (ESVs) to create a sample x ESV table that contains read counts. Arrow 2 indicates where pseudogenes can be removed using two different approaches. The first method translates ESVs, retains the longest nucleotide open reading frame (ORF), then removes sequences with very small or very large outlier lengths. The second method retains the amino acid sequence from the longest ORF, does a profile HMM analysis, then removes sequences with very small outlier full sequence bit scores. Arrow 3 indicates where rare sequence clusters from each sample are removed and read numbers are mapped to the final report. The final report contains all ESVs for each sample, read numbers, ORF sequences, and taxonomic assignments with bootstrap support values