Abstract
PURPOSE
We aimed to explore whether multiparametric magnetic resonance imaging (MRI)-based radiomics combined with selected blood inflammatory markers could effectively predict the grade and proliferation in glioma patients.
METHODS
This retrospective study included 152 patients histopathologically diagnosed with glioma. Stratified sampling was used to divide all patients into a training cohort (n=107) and a validation cohort (n=45) according to a ratio of 7:3, and five-fold repeat cross-validation was adopted in the training cohort. Multiparametric MRI and clinical parameters, including age, the neutrophil-lymphocyte ratio and red cell distribution width, were assessed. During image processing, image registration and gray normalization were conducted. A radiomics analysis was performed by extracting 1584 multiparametric MRI-based features, and the least absolute shrinkage and selection operator (LASSO) was applied to generate a radiomics signature for predicting grade and Ki-67 index in both training and validation cohorts. Statistical analysis included analysis of variance, Pearson correlation, intraclass correlation coefficient, multivariate logistic regression, Hosmer–Lemeshow test, and receiver operating characteristic (ROC) curve.
RESULTS
The radiomics signature demonstrated good performance in both the training and validation cohorts, with areas under the ROC curve (AUCs) of 0.92, 0.91, and 0.94 and 0.94, 0.75, and 0.82 for differentiating between low and high grade gliomas, grade III and grade IV gliomas, and low Ki-67 and high Ki-67, respectively, and was better than the clinical model; the AUCs of the combined model were 0.93, 0.91, and 0.95 and 0.94, 0.76, and 0.80, respectively.
CONCLUSION
Both the radiomics signature and combined model showed high diagnostic efficacy and outperformed the clinical model. The clinical factors did not provide additional improvement in the prediction of the grade and proliferation index in glioma patients, but the stability was improved.
Glioma is the most common primary malignant tumor in the brain, accounting for nearly 81% of malignant tumors in the brain (1). In the 2016 World Health Organization (WHO) classification system, gliomas can be graded from level I to level IV according to the histopathological and clinical criteria. Low-grade gliomas (LGGs) include grades I and II, whereas high-grade gliomas (HGGs) include grades III and IV (2, 3). Patients with LGGs may remain indolent for years or progress to glioblastoma (4) and are generally treated with only surgery and in some cases with radiotherapy (5). For patients with HGGs, radiotherapy and/or chemotherapy are usually followed after surgery, which often indicates a poor prognosis (6). Although genetic factors play an increasingly important role in indicating prognosis and survival (2, 3, 7), false classification could directly cause inappropriate treatment, which significantly affects patient outcomes. Hence, precise grading is still crucial for therapy planning and progression assessment in patients with gliomas.
The nuclear protein Ki-67 is a reliable biomarker for evaluating the proliferation of tumor cells, because it only exists in active phase of the cell cycle. (8). The Ki-67 labeling index (LI) is defined as the percentage of Ki-67-positive tumor nuclei of all tumor nuclei (9). The WHO reported that the Ki-67 LI values are below 4% of the mean value for grade II glioma, 5%–10% for grade III glioma, and 15%–20% for grade IV glioma (2). The overexpression of Ki-67 infers poor differentiation and prognosis (10). Hence, the accurate preoperative evaluation of the Ki-67 LI maybe helpful in treatment decision-making for glioma patients.
Recently, there is a newly emerging area in radiology: radiomics, which is realized by quantifying image data with advanced image post-processing techniques (11). For tumor radiomics, a complete characterization of the tumor is established by feature selection and high-throughput analysis (12, 13). The radiomics analysis of glioma MRI could predict the grade and proliferation potency in glioma patients (14–16). A study showed that multiparametric MRI-based radiomics analysis, assisted by T2-weighted imaging, gadolinium-based contrast-enhanced T1-weighted imaging and the apparent diffusion coefficient (ADC), could help clinicians classify gliomas more accurately (17).
Chronic inflammation, which is part of the tumor microenvironment, is associated with tumor occurrence and progression (18, 19). Some blood inflammatory biomarkers, such as the neutrophil-lymphocyte ratio (NLR), platelet count, and red cell distribution width (RDW), have been proposed to predict the prognosis of tumors (20–22). And recent studies showed that the NLR and platelet count in glioblastoma patients (23, 24) and the NLR and RDW in glioma patients were significant in predicting prognosis (25).
To our knowledge, there have been no studies on MRI-based radiomics combined with selected blood inflammatory markers to predict the grade and proliferation potency in glioma patients. The purpose of this study is to build and assess a radiomics nomogram to preoperatively predict glioma grade and proliferation potency by incorporating MRI-based radiomics and clinical signatures, including age, NLR, and RDW.
Methods
The radiological and surgical databases were retrospectively reviewed for records from 2013 to 2018. This study was approved by our institutional ethics committee and the requirement for informed consent was waived due to retrospective analysis (202001). The inclusion criteria for patients were: (a) Histopathologically confirmed supratentorial glioma except grade I; (b) Preoperative craniocerebral MRI including T2- and T1-weighted imaging, T2-weighted attenuated inversion recovery imaging (T2 FLAIR) and contrast-enhanced T1-weighted imaging; and (c) patients with Ki-67 LI values. Nineteen of these patients were excluded because of: treatment (chemoradiotherapy, chemotherapy or radiotherapy) prior to the MRI examination (n=10); and poor imaging quality rendering the image unqualified for image analysis due to the influence of motion or susceptibility (n=9). Finally, this study included 152 patients (47, 39 and 66 patients were diagnosed as grade II, III and IV gliomas, respectively). In our study, the low expression level of Ki-67 was defined as ≤10% positive staining (n=56), whereas the high expression level of Ki-67 was defined as >10% positive staining (n=96) (26). Stratified sampling was used to divide all patients into a training cohort (n=107) and a validation cohort (n=45) according to a ratio of 7:3.
MRI protocol
All included patients were examined with a 3.0 Tesla MRI scanner (Trio, Siemens). Axial T2-weighted imaging, T2 FLAIR, unenhanced and contrast-enhanced T1-weighted imaging were performed. The acquisition parameters were as follows: T2-weighted imaging: slice thickness, 4.5 mm; intersection gap, 4.5 mm; repetition time (TR)/echo time (TE), 7140/98 ms; field of view (FOV), 22×22 cm2; pixel matrix, 384×384; acquisition time, 2 min 17 s; flip angle, 150°; T2 FLAIR: slice thickness, 4.5 mm; intersection gap, 0.45 mm; TR/TE, 8400/91 ms; FOV, 24×24 cm2; pixel matrix, 256×256; acquisition time, 2 min 16 s; flip angle, 130°; and T1-weighted imaging: slice thickness, 4.5 mm; intersection gap, 0.45 mm; TR/TE, 220/3 ms; FOV, 22×22 cm2; pixel matrix, 256×256; acquisition time, 44 s; flip angle, 70°. A dosage of 0.2 mL/kg gadopentetate dimeglumine was given intravenously and axial contrast-enhanced T1-weighted imaging was acquired.
Volume-of-interest (VOI) segmentation
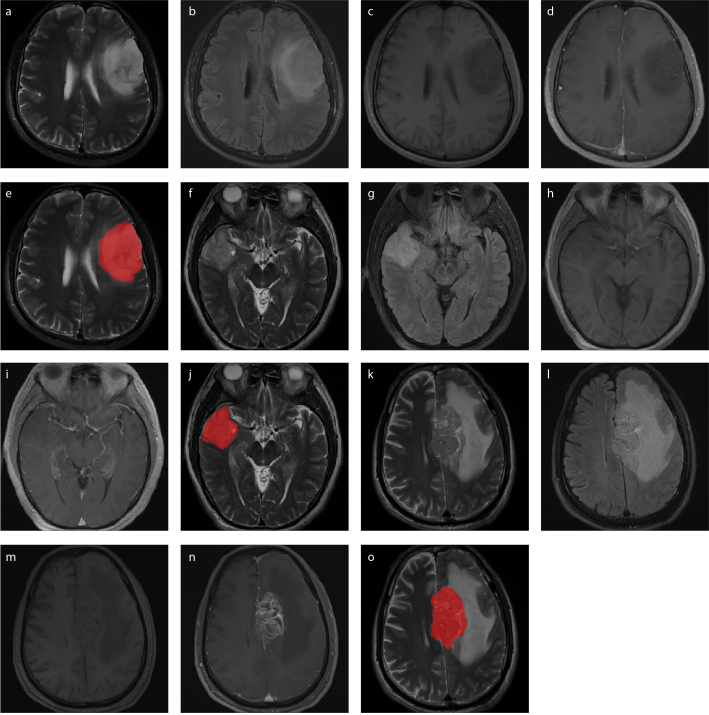
The T2 FLAIR, unenhanced and contrast-enhanced T1-weighted imaging were co-registered with reference to the T2-weighted imaging using Slicer (version: 4.10.1, package: Elastix, https://www.slicer.org). The tumor VOIs covered the most representative regions of the gliomas by manual segmentation in the T2-weighted imaging using the ITK-SNAP software (version: 3.8.0, https://www.itksnap.org). For the VOI delineation, the tumor boundaries of nonenhancing gliomas were determined in T2-weighted imaging, because in these tumors hyperintense signals represented the tumor regions of gliomas; meanwhile, for the enhancing gliomas the most representative regions were decided in T2-weighted imaging with reference to the contrast-enhanced T1-weighted imaging to separate the parenchymal areas from edematous areas and avoid the inclusion of obvious cysts, necrosis and hemorrhage regions. Then, the contours of the VOIs on the T2-weighted imaging were copied to images of other MRI sequences. The segmentation of the VOIs was performed independently by a senior neuroradiologist. To verify the consistency between readers, the VOIs of 30 randomly selected cases were segmented by another senior neuroradiologist. The segmentation of the VOIs is presented in Fig. 1.
Figure 1. a–o.
Image acquisition and volume-of-interest (VOI) segmentation. T2-weighted images (a, f, k), T2-weighted attenuated inversion recovery images (b, g, l), T1-weighted images (c, h, m), contrast-enhanced T1-weighted images (d, i, n) and tumor region (red in T2-weighted images) (e, j, o) show VOI segmentation in patients with grade II (a–e), grade III (f–j), and grade IV (k–o) glioma.
Radiomics features extraction and radiomics signature construction
All MRI data underwent image resampling to isotropic voxels (1×1×1 mm) with linear interpolate, and intensity normalization (Z-score) before feature extraction (27) and radiomics features were calculated by Artificial Intelligence Kit (AK, version: 3.2.1, GE Healthcare). Overall, 396 radiomics features were extracted from each sequence for each patient, including 42 histogram features, 9 form factor features, 144 gray level cooccurrence matrix (GLCM) features, 180 run-length matrix (RLM) features, 11 gray level size zone matrix (GLSZM) features and 10 Haralick features.
The interobserver intraclass correlation coefficient (ICC) selects values greater than 0.75, and T2 FLAIR, unenhanced and contrast-enhanced T1-weighted imaging and T2-weighted imaging had 346, 358, 357, and 243 features reserved, respectively. The results indicated favorable interobserver feature extraction reproducibility. Before feature selection, all the radiomics features were standardized by the Z-score method. Then, feature selection was conducted as follows: First, analysis of variance (ANOVA) was performed to remove features with p > 0.05, followed by the correlation test to remove features that had a correlation coefficient > 0.9. Next, the least absolute shrinkage and selection operator (LASSO) model, which could improve prediction accuracy and interpretation (28, 29), was used to further select the features. The reserved features were applied to construct the radiomics score for every patient.
Assessment of the radiomics score, clinical model and radiomics nomogram
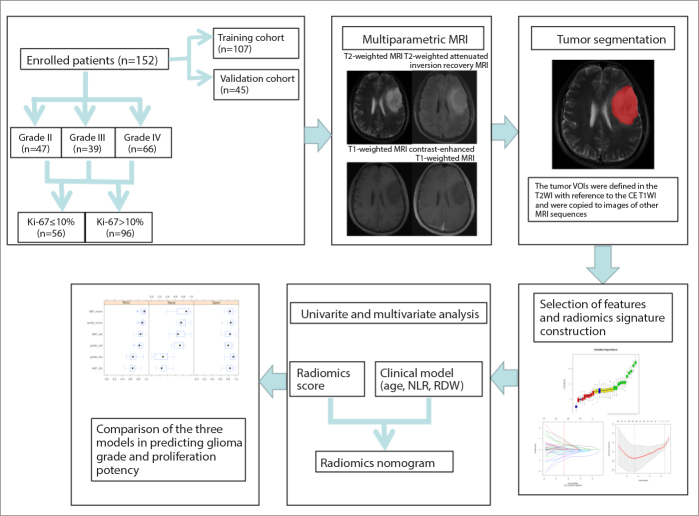
The radiomics score, which was derived from individual sequences and multiparametric MRI, and significant clinical data (age, NLR, and RDW) (23–26) were used as potential predictors for the multivariate logistic regression analysis to build a prediction model of the grade and proliferation potency of gliomas. The radiomics nomogram was also constructed by combining the radiomics score derived from multiparametric MRI and clinical factors. The performance of the radiomics score, clinical model and radiomics nomogram was evaluated by using a calibration curve. The radiomics score, clinical model and radiomics nomogram were applied in the validation cohort subsequently by using the same formula in the training cohort. Additionally, the no-information rate was calculated in our dataset for comparison with the diagnosis accuracy. A simplified flowchart of the study is given in Fig. 2.
Figure 2.
Flowchart of constructing a radiomic nomogram with multiparametric MRI and clinical features to predict the glioma grade and proliferation potency. T2WI, T2-weighted imaging; CE T1WI, contrast-enhanced T1-weighted imaging; NLR, neutrophil-lymphocyte ratio; RDW, red cell distribution width.
Statistical analysis
The LASSO method was utilized in conjunction with the penalty parameter adjustment and is performed through five-fold repeat cross-validation based on the minimum deviation criteria. Backward stepwise selection was utilized by dint of the likelihood ratio test, and Akaike’s information criterion (AIC) was taken as the stopping rule for the multivariate logistic regression.
The accuracy of the radiomics score was quantified by the area under the receiver operating characteristic (ROC) curve (AUC) in both the training and validation cohorts. The range of 95% confidence interval was used to evaluate the stability of the models. The Hosmer–Lemeshow test were used to assess the goodness-of-fit and discriminatory ability. ICCs were used to evaluate the interobserver agreement.
All the statistical analyses were conducted with R software (version 3.5.3; https://www.r-project.org). The “rms” package was used to construct the nomogram. All the reported statistical significance levels were two-sided, with statistical significance of 0.05.
Results
There were no significant differences in age, NLR, RDW, glioma grading, and Ki-67 expression (p > 0.05) between the training and validation cohorts. There were statistically significant differences in age and NLR variables among LGGs and HGGs. The characteristics of all the participants are shown in Table 1 and Supplementary Tables 1–3.
Table 1.
Characteristics of the patients in the training and validation cohorts
| Training cohort (n=107) | Validation cohort (n=45) | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Grade II | Grade III | Grade IV | Grade II | Grade III | Grade IV | ||
| n | 36 | 28 | 43 | 11 | 11 | 23 | |
|
| |||||||
| Age (years), mean±SD | 44.38±11.31 | 48.04±12.02 | 54.46±12.58 | 40.18±8.85 | 56.73±10.98 | 56.83±13.07 | |
|
| |||||||
| NLR, median (Q1, Q3) | 2.07 (1.72, 3.22) | 2.44 (1.84, 3.51) | 2.83 (2.07, 5.14) | 1.83 (1.48, 2.9) | 3.42 (2.56, 4.4) | 3.00 (2.30, 5.4) | |
|
| |||||||
| RDW, mean±SD | 13.1±1.04 | 13.34±1.05 | 13.37±1.52 | 13.21±1.38 | 12.87±0.58 | 13.42±0.86 | |
|
| |||||||
| Ki-67 LI, n | ≤10% | 42 | 14 | ||||
| >10% | 65 | 31 | |||||
SD, standard deviation; NLR, neutrophil-lymphocyte ratio; Q1–Q3, 25th and 75th percentiles; RDW, red cell distribution width; Ki-67 LI, Ki-67 labeling index.
Table 3.
Diagnostic performance of the three proposed models
| Different models | Training cohort | Validation cohort | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Accuracy | AUC (95% CI) | Sensitivity | Specificity | Accuracy | AUC (95% CI) | ||
| Clinical | LGGs and HGGs | 0.80 | 0.52 | 0.62 | 0.70 (0.59–0.80) | 0.64 | 0.97 | 0.89 | 0.82 (0.66–0.99) |
| Grade III and Grade IV | 0.82 | 0.57 | 0.66 | 0.65 (0.51–0.78) | 1.00 | 0.30 | 0.55 | 0.60 (0.39–0.80) | |
| Low Ki-67 LI and High Ki-67 LI | 0.45 | 0.82 | 0.67 | 0.67 (0.56–0.77) | 0.43 | 1.00 | 0.82 | 0.72 (0.54–0.90) | |
|
| |||||||||
| Radiomics | LGGs and HGGs | 0.94 | 0.85 | 0.88 | 0.92 (0.87–0.97) | 1.00 | 0.82 | 0.87 | 0.94 (0.87–1.00) |
| Grade III and Grade IV | 0.86 | 0.89 | 0.88 | 0.91 (0.83–0.99) | 0.82 | 0.60 | 0.68 | 0.75 (0.57–0.93) | |
| Low Ki-67 LI and High Ki-67 LI | 0.93 | 0.83 | 0.87 | 0.94 (0.89–0.98) | 0.64 | 0.94 | 0.84 | 0.82 (0.69–0.96) | |
|
| |||||||||
| Combined | LGGs and HGGs | 0.92 | 0.85 | 0.87 | 0.93 (0.89–0.98) | 1.00 | 0.88 | 0.91 | 0.94 (0.87–1.00) |
| Grade III and Grade IV | 0.79 | 0.96 | 0.89 | 0.91 (0.83–0.99) | 0.73 | 0.70 | 0.80 | 0.76 (0.58–0.94) | |
| Low Ki-67 LI and High Ki-67 LI | 0.91 | 0.88 | 0.89 | 0.95 (0.91–0.99) | 0.86 | 0.68 | 0.73 | 0.80 (0.66–0.94) | |
AUC, area under the curve; 95% CI, 95% confidence interval; LGGs, low-grade gliomas; HGGs, high-grade gliomas; Ki-67 LI, Ki-67 labeling index.
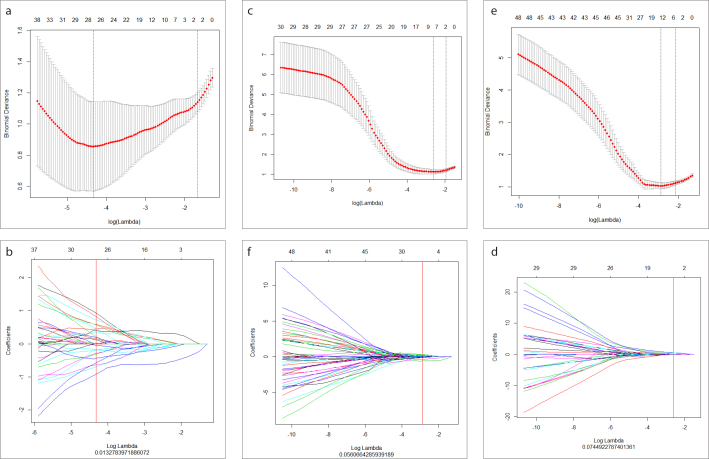
The results of the data dimension reduction with LASSO are shown in Fig. 3a, 3c, 3e. LASSO with 11, 9, and 12 of the most important variables and their coefficients are shown in Fig. 3b, 3d, 3f, which indicate great diagnostic potential. All the reserved features and the best-performing features are listed in Table 2. The logistic regression models predicted the grade and proliferation potency of gliomas as shown in the following formulas:
Figure 3. a–f.
Feature selection using LASSO. The tuning parameter (λ) selection in the LASSO model used ten-fold cross-validation with minimum criteria and could predict low-grade gliomas (LGGs) and high-grade gliomas (HGGs) (a), grade III and grade IV gliomas (c), and low Ki-67 and high Ki-67 (e). LASSO coefficient profiles of texture features could predict LGGs and HGGs (b), grade III and grade IV gliomas (d), and low Ki-67 and high Ki-67 (f).
Table 2.
Radiomic feature selection results
| T2WI | T1WI | T2 FLAIR | CE T1WI | |
|---|---|---|---|---|
| LGGs and HGGs | 1 | 1 | 3 | |
| Skewness | Inverse DifferenceMoment_angle45_offset7 | Kurtosis | ||
| Inertia_angle90_ offset1 uniformity | ||||
|
| ||||
| Grade III and IV | 2 | 1 | 5 | |
| Percentile95 | MinIntensity | kurtosis | ||
| Range | SmallAreaEmphasis | |||
| LowIntensity SmallAreaEmphasis | ||||
| Variance | ||||
| Inertia_angle90_ offset1 | ||||
|
| ||||
| Low Ki-67 LI and high Ki-67 LI | 2 | 1 | 1 | 3 |
| Skewness | t1_LongRunEmphasis_angle45_offset1 | flair_Correlation_angle0_offset7 | t1c_kurtosis | |
| ShortRunHighGreyLevelEmphasis_AllDirection_offset4_SD | Inertia_angle90_offset1 | |||
| t1c_LargeAreaEmphasis | ||||
T2WI, T2-weighted imaging; T1WI, T1-weighted imaging; T2 FLAIR, T2-weighted attenuated inversion recovery imaging; CE T1WI, contrast-enhanced T1-weighted imaging; LGGs, low-grade gliomas; HGGs, high-grade gliomas; Ki-67 LI, Ki-67 labeling index.
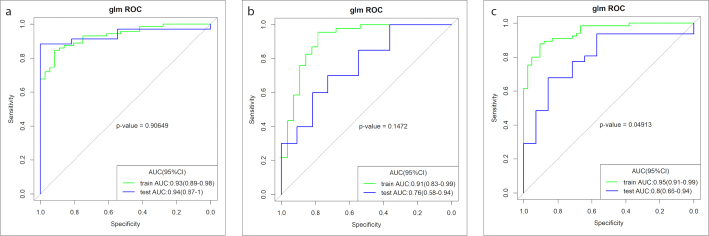
The AUCs of the radiomics score derived from multiparametric MRI were significantly higher than those of the models of individual sequences in differentiating between LGGs and HGGs, grade III and grade IV glioma, and low Ki-67 and high Ki-67 in the training and validation cohorts (Supplementary Fig.). The radiomics score derived from multiparametric MRI was significantly correlated with glioma grading and Ki-67 expression (p < 0.001). In the training cohort, the AUCs of the radiomics score for differentiating between LGGs and HGGs, grade III and grade IV glioma, and low Ki-67 and high Ki-67 were 0.92, 0.91 and 0.94, respectively, which were significantly higher than those of clinical factors, and the De Long test p values were all < 0.001. After combining the radiomics score and clinical factors, the AUCs were 0.93, 0.91 and 0.95, respectively (Fig. 4), and all of the De Long test p values were < 0.001 compared with clinical factors. These results suggested that the performance was best when multiparametric MRI and clinical factors were considered simultaneously.
Figure 4. a–c.
The ROC curves show the effectiveness of combining the radiomics score and clinical factors in differentiating between LGGs and HGGs (a), grade III and grade IV gliomas (b), and low Ki-67 and high Ki-67 (c) in the training cohort.
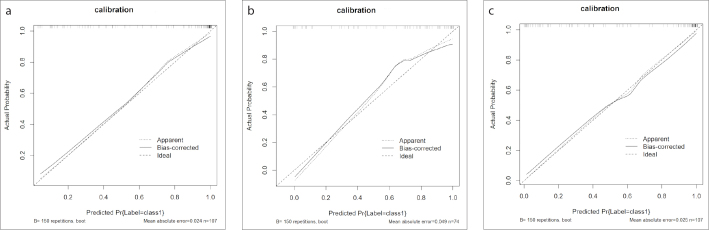
The calibration curve of radiomics nomogram also showed that the predicted probability was in good agreement with actual probability in the training cohort (Fig. 5). The predictive performances of the radiomics score, clinical factors and their combination are shown in Fig. 6. The Hosmer–Lemeshow test demonstrated no significant differences (p = 1.00, p = 0.39, and p = 0.24) and indicated no deviation from a perfect fit.
Figure 5. a–c.
Calibration of the radiomics nomogram to predict LGGs and HGGs (a), grade III and grade IV gliomas (b), and low Ki-67 and high Ki-67 (c) in the training cohort.
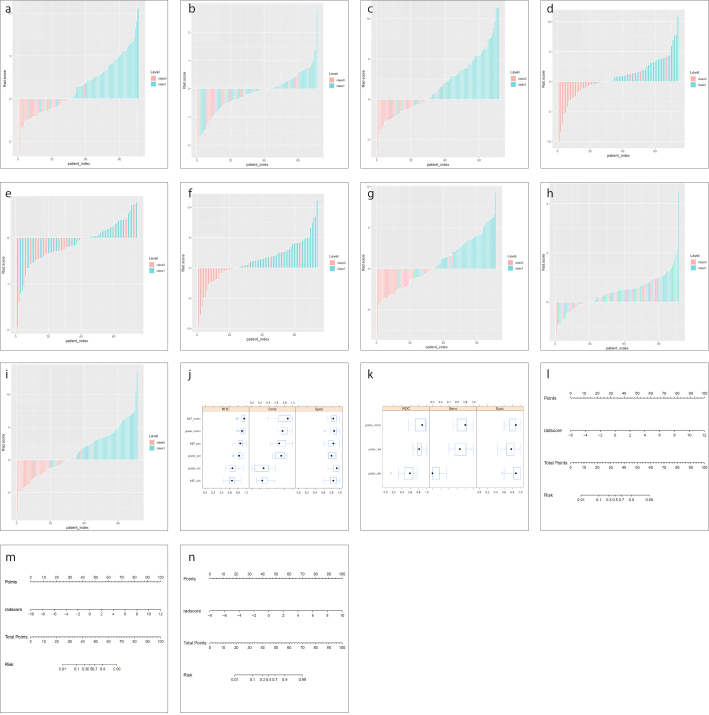
Figure 6. a–n.
The predictive performance of the radiomics score, clinical factors and their combination. The bar plots for the training cohort show the radiomics scores, clinical factors and their combination for each patient categorized by LGGs and HGGs (a–c), grade III and grade IV gliomas (d–f), and low Ki-67 and high Ki-67 (g–i). The boxplots show the radiomics scores, clinical factors and their combination in training cohort categorized by LGGs and HGGs (j), low Ki-67 and high Ki-67 (j), and grade III and grade IV gliomas (k). The radiomics nomogram developed in the training cohort with the radiomic score, age and red cell distribution width (RDW) of every patient to detect LGGs and HGGs (l), grade III and grade IV gliomas (m), and low Ki-67 and high Ki-67 (n).
In the validation cohort, the findings were similar. The radiomics score and combination strategy showed favorable performances. The Hosmer–Lemeshow test indicated no significant difference (p = 1.00) in differentiating between LGGs and HGGs, but significant differences in differentiating between grade III and grade IV gliomas (p = 0.002) and low Ki-67 and high Ki-67 (p = 0.002). The diagnostic performance of the three proposed models is shown in Table 3. Additionally, for the measure of class imbalance, the no-information rates between LGGs and HGGs, grade III and grade IV glioma, and low and high Ki-67 were 0.6216, 0.621 and 0.6075 in the training cohort and 0.645, 0.645 and 0.689 in the validation cohort, respectively. The radiomics and nomogram model prediction accuracy all exceeded the no-information rate.
Discussion
This study predicted the grade and proliferation potency in glioma patients by developing a radiomics nomogram. The model combined multiparametric MRI-based radiomics and clinical factors, including age and selected blood inflammatory markers, and showed good discriminative ability.
The conventional presurgical determination of glioma grades mainly depends on conventional MRI techniques, including T2- and T1-weighted imaging, T2 FLAIR and, particularly, contrast-enhanced T1-weighted imaging. However, conventional MRI provides limited information regarding glioma grading, and the accuracy is only 55% to 83%. A study showed that 20% of LGGs show contrast enhancement, whereas a third of HGGs do not show contrast enhancement (30). In addition, conventional MRI is unable to evaluate tumor proliferation potency. A few studies have used advanced MRI sequences, such as multi-b-value diffusion-weighted imaging, diffusion tensor imaging (DTI), diffusion kurtosis imaging (DKI), and dynamic contrast-enhanced perfusion imaging (DCE-MRI), for the grading or proliferation potency evaluation of gliomas (31–37). Ren et al. (31) showed that the heterogeneity index α and slow diffusion coefficient derived from non-Gaussian diffusion MRI could improve the accuracy of glioma grading and predict the proliferation level of malignant gliomas. Mean diffusivity derived from DTI could also be used in grading gliomas, and the ROC curve showed a sensitivity of 91.8% (32). It has also been reported that DKI is superior to DTI in predicting the grade and proliferation potency, and average kurtosis is suggested to be the best prediction parameter (33–36). Jain et al. (37) demonstrated some potential advanced MRI parameters for differentiating LGGs and HGGs. The sensitivity and specificity of the relative cerebral blood volume (rCBV) in differentiating LGGs and HGGs were 97.22% and 100%, respectively. Additionally, rCBV, volume transfer constant (Ktrans) and volume of extravascular extracellular space per unit volume of tissue (Ve) were associated with the Ki-67 LI in HGGs (38). Data analysis based on advanced imaging has gained increasing attention; however, the time-consuming processes and some uncertain results limit its fast popularization.
The radiomics score was an independent predictor of the grade and proliferation potency in glioma patients (39). Wang et al. (17) extracted features from T2-weighted imaging, ADC and contrast-enhanced T1-weighted imaging sequences. They compared the radiomic features of HGGs with that of LGGs. Their results showed 15 features with significant differences between HGGs and LGGs, and the diagnostic efficiency could be improved by combining these 15 features (with AUCs of 0.971 and 0.961 in the training and validation cohorts, respectively). This approach indicates that multiparametric MRI-based radiomic features play an important role in glioma grading. Cho et al. (40) combined T2-weighted imaging, T1-weighted imaging, T2 FLAIR and contrast-enhanced T1-weighted imaging and used machine learning, feature selection techniques and a radiomics approach for glioma grading. They extracted 5 significant features that showed an average AUC of 0.9400 for the training cohort and 0.9030 for the validation cohort.
In our study, we performed a radiomics analysis based on individual sequences and multiparametric MRI, including T2-weighted imaging, T1-weighted imaging, T2 FLAIR and contrast-enhanced T1-weighted imaging. The results showed that the radiomics score derived from multiparametric MRI was superior to that of the models of individual sequences, and in the former the results showed that the selected radiomics features related to heterogeneities in T2 FLAIR and contrast-enhanced T1-weighted imaging were the most important components in differentiating LGGs and HGGs, grade III and grade IV, and low and high Ki-67. The reason might be because T2 FLAIR could provide more information on edema, and contrast-enhanced T1-weighted imaging contains more information about blood supply, necrosis and blood-brain barrier damage due to gliomas, which are related with the grading and Ki-67 LI of gliomas. Skewness, kurtosis, range, variance, uniformity and percentil95 are first order features used to describe the degree of uniform distribution of signal strength in tumors. Inertia, InverseDifferenceMoment, ClusterProminence, ShortRunHighGrayLevelEmphasis and LongRunEmphasis are texture features that mainly describe the heterogeneity within the tumor, and the heterogeneity within malignant tumors is higher. The AUCs of the radiomics score were 0.92 and 0.94 in the training cohort and validation cohort for differentiating LGGs and HGGs, respectively, which is consistent with that of a previous study (14). In addition, grade III and grade IV gliomas could be well differentiated by the extracted variables with LASSO, and the AUC was 0.91 and 0.75 in the training cohort and validation cohort, which is lower than that for the differentiation of LGGs and HGGs. However, the performance is of value in differentiating grade III and grade IV gliomas. Due to the differences of prognosis and treatment between grade III and grade IV gliomas, their classification is essential. Even though only a few studies have focused on differentiating grade III and grade IV gliomas, our results were consistent with those of previous studies (AUCs of 0.881 and 0.992) (16, 39). However, those studies were limited by the lack of an independent validation cohort.
A previous study investigated the radiomic features related to the Ki-67 LI in gliomas for tumor proliferation prediction. The results showed that the radiomics signature based on T2-weighted imaging could predict Ki-67 LI in LGGs, with accuracies of 83.3% and 88.6% (AUCs of 0.91 and 0.93) in the training and validation cohorts, respectively (40). Su et al. (16) explored the diagnostic performance of radiomics based on multiparametric MRI, including T1- and T2-weighted imaging, T2 FLAIR, contrast-enhanced T1-weighted imaging, ADC and arterial spin labeling, for predicting tumor proliferation and found that the AUC was 0.936, which is consistent with our results (AUCs of 0.94 and 0.82 in the training and validation cohort, respectively). Hence, multiparametric MRI-based radiomics could be used to predict the proliferation potency of gliomas.
Chronic inflammation is characteristic of tumor microenvironment, stimulating tumor progression and metastasis. Some studies have suggested that peripheral inflammatory states, such as the NLR, platelet count and RDW, could predict prognosis in patients with glioma (23, 24). Auezova et al. (25) assessed the association of the NLR, platelet count and RDW with tumor grade and survival in glioma patients and found that only the NLR and RDW was helpful to assess progression and outcomes. Weng et al. (41) suggested that the combination of radiologic biomarkers (including contrast enhancement) and peripheral parameters such as the NLR could successfully predict proliferation potency in glioma patients. They found that there were significant correlations between the NLR and tumor grade or proliferation potency and between the platelet count and tumor grade or proliferation potency, whereas no correlation existed between the monocyte-lymphocyte ratio (MLR) and grade or proliferation potency. However, they assessed the value of features from only conventional MRI such as contrast enhancement, and radiomics based on multiparametric MRI was not included. In our study, age, NLR and RDW were selected to evaluate the grade and proliferation potency in glioma patients. The combination of these three clinical factors was associated with the grade (AUCs for differentiating between LGGs and HGGs of 0.7 and 0.82 in the training and validation cohort, respectively; AUCs for differentiating between grade III and grade IV gliomas of 0.65 and 0.60 in the training and validation cohort, respectively) and proliferation potency (AUCs of 0.67 and 0.72 in the training and validation cohort, respectively). Nevertheless, the performance was significantly weaker than that of the radiomics score.
This study has several limitations. First, this is a retrospective evaluation with the data collected from a single center. Multicenter studies with greater sample sizes are needed. Second, the advanced MRI sequences (such as DWI, DTI or perfusion imaging) were not used in this study and need to be added in the future. Third, the 2016 WHO classification includes molecular characteristics such as IDH mutation and 1p/19q codeletion, but these factors were not taken into account in our study. Therefore, molecular markers should be considered in future studies. Fourth, the sample size was relatively limited, and only 39 patients had grade III gliomas. Therefore, the calibration in the validation cohort was not optimal. Finally, the prognostic role of the radiomics score associated with inflammation markers was not assessed and needs to be validated in the future.
In conclusion, multiparametric MRI-based radiomics and peripheral inflammatory markers (age, NLR, and RDW) can be used to assess the grade and proliferation potency accurately for gliomas preoperatively and noninvasively. Such a method is valuable in designing personalized treatment strategies.
Main points.
The clinical model included age, neutrophil-lymphocyte ratio and red cell distribution width.
Radiomics analysis is based on multiparametric MRI, including T2-weighted, T1-weighted, T2 FLAIR and contrast-enhanced T1-weighted imaging.
Among the radiomics features, factors related to heterogeneities in T2 FLAIR and contrast-enhanced T1-weighted imaging were the most important components for predicting between LGGs and HGGs, grade III and grade IV, and low Ki-67 and high Ki-67.
Radiomics was better for differentiating LGGs and HGGs, grade III and grade IV gliomas, and low Ki-67 and high Ki-67 than the clinical model.
The combination of radiomics and the clinical model could improve the predictive stability of the grade and proliferation in glioma patients.
Supplementary Information
Supplementary Table 1.
Characteristics of LGGs and HGGs patients in the training and validation cohorts
| Training cohort (n=107) | Validation cohort (n=45) | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| LGGs | HGGs | p | LGGs | HGGs | p | |
| Number | 36 | 71 | - | 11 | 34 | - |
|
| ||||||
| Age | ||||||
| Mean±SD | 44.53±11.31 | 51.93±22.68 | 0.001 | 40.18±8.85 | 56.79±12.27 | <0.001 |
|
| ||||||
| NLR | ||||||
| Median (Q1, Q3) | 2.07 (1.72, 3.21) | 2.73 (1.97, 4.64) | 0.036 | 1.83 (1.47, 2.89) | 3.21 (2.28, 5.12) | 0.016 |
|
| ||||||
| RDW | ||||||
| Mean±SD | 13.1±1.04 | 13.36±1.34 | 0.161 | 13.21±1.38 | 13.24±0.82 | 0.283 |
p < 0.05.
NLR, neutrophil-lymphocyte ratio; RDW, red cell distribution width; LGGs, low-grade gliomas; HGGs, high-grade gliomas.
Supplementary Table 2.
Characteristics of Grade III and IV patients in the training and validation cohorts
| Training cohort (n=74) | Validation cohort (n=31) | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Grade III | Grade IV | p | Grade III | Grade IV | p | |
| Number | 28 | 46 | - | 11 | 20 | |
|
| ||||||
| Age | ||||||
| Mean±SD | 50.18±13.30 | 54.72±12.44 | 0.099 | 51.27±13.52 | 26.6±13.25 | 0.082 |
|
| ||||||
| NLR | ||||||
| Median (Q1, Q3) | 3.04 (2.18, 4.63) | 2.8 (2.07, 4.72) | 0.608 | 2.25 (2.18, 2.6) | 3.20 (2.35, 6.42) | 0.043 |
|
| ||||||
| RDW | ||||||
| Mean±SD | 13.18±1.05 | 13.26±0.92 | 0.448 | 13.28±0.71 | 13.69±1.94 | 0.885 |
p < 0.05.
NLR, neutrophil-lymphocyte ratio; RDW, red cell distribution width.
Supplementary Table 3.
Characteristics of low Ki-67 and high Ki-67 patients in the training and validation cohorts
| Training cohort (n=107) | Validation cohort (n=45) | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Low Ki-67 LI | High Ki-67 LI | p | Low Ki-67 LI | High Ki-67 LI | p | |
| Number | 42 | 65 | - | 14 | 31 | - |
|
| ||||||
| Age | ||||||
| Mean±SD | 46.4±12.42 | 51.6±12.48 | 0.018 | 42.86±9.65 | 57.19±12.73 | 0.001 |
|
| ||||||
| NLR | ||||||
| Median (Q1, Q3) | 2.22 (1.75, 3.48) | 2.51 (2.07, 4.53) | 0.127 | 2.2 (2.34, 5.4) | 3.42 (2.34, 5.4) | 0.013 |
|
| ||||||
| RDW | ||||||
| Mean±SD | 13.01±0.84 | 13.44±13.0 | 0.116 | 13.17±1.24 | 13.26±0.84 | 0.233 |
p < 0.05.
NLR, neutrophil-lymphocyte ratio; RDW, red cell distribution width; Ki-67 LI, Ki-67 labeling index.
Compared with the individual sequences, the ROC curves showed the effectiveness of the multiparametric MRI in differentiating between LGGs and HGGs, grade III and grade IV gliomas, and low Ki-67 and high Ki-67 in the training cohort (a–c) and validation cohort (d–f), respectively.
Footnotes
Conflict of interest disclosure
The authors declared no conflicts of interest.
References
- 1.Ostrom QT, Bauchet L, Davis FG, et al. The epidemiology of glioma in adults: a “state of the science” review. Neuro Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Louis DN, Ohgaki H, Wiestler OD, et al. WHO classification of Tumours of the central nervous system. 4th Ed revised. Lyon: IARC; 2016. [Google Scholar]
- 3.Gupta A, Dwivedi T. A simplified overview of World Health Organization Classification Update of Central Nervous System Tumors 2016. J Neurosci Rural Pract. 2017;8:629–641. doi: 10.4103/jnrp.jnrp_168_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brat DJ, Verhaak RG, Aldape KD, et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Rhun E, Taillibert S, Chamberlain MC. Current management of adult diffuse infiltrative low grade gliomas. Curr Neurol Neurosci Rep. 2016;16:15. doi: 10.1007/s11910-015-0615-4. [DOI] [PubMed] [Google Scholar]
- 6.Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 7.Ren Y, Zhang X, Rui WT, et al. Noninvasive prediction of IDH1 mutation and ATRX expression loss in low-grade gliomas using multiparametric MR radiomic features. J Magn Reson Imaging. 2019;49:808–817. doi: 10.1002/jmri.26240. [DOI] [PubMed] [Google Scholar]
- 8.Scholzen T, Gerdes J. The Ki-67 protein: from the known and th. J Cell Physiol. 2000;182:311–322. doi: 10.1002/(SICI)1097-4652(200003)182:3<311::AID-JCP1>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 9.Johannessen AL, Torp SH. The clinical value of Ki-67/MIB-1 labeling index in human astrocytomas. Pathol Oncol Res. 2006;12:143–147. doi: 10.1007/BF02893360. [DOI] [PubMed] [Google Scholar]
- 10.Chen WJ, He DS, Tang RX, et al. Ki-67 is a valuable prognostic factor in gliomas: evidence from a systematic review and meta-analysis. Asian Pac J Cancer Prev. 2015;16:411–420. doi: 10.7314/APJCP.2015.16.2.411. [DOI] [PubMed] [Google Scholar]
- 11.Yip SS, Aerts HJ. Applications and limitations of radiomics. Phys Med Biol. 2016;61:R150–166. doi: 10.1088/0031-9155/61/13/R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Xu X, Tian Q, et al. Radiomics assessment of bladder cancer grade using texture features from diffusion-weighted imaging. J Magn Reson Imaging. 2017;46:1281–1288. doi: 10.1002/jmri.25669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fehr D, Veeraraghavan H, Wibmer A, et al. Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images. Proc Natl Acad Sci USA. 2015;112:e6265–6273. doi: 10.1073/pnas.1505935112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho HH, Lee SH, Kim J, et al. Classification of the glioma grading using radiomics analysis. Peerj. 2018;6:e5982. doi: 10.7717/peerj.5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwanho C, Hyunjin P. Classification of low-grade and high-grade glioma using multi-modal image radiomics features. Conf Proc IEEE Eng Med Biol Soc; 2017; pp. 3081–3084. [DOI] [PubMed] [Google Scholar]
- 16.Su CL, Jiang JJ, Zhang S, et al. Radiomics based on multicontrast MRI can precisely differentiate among glioma subtypes and predict tumour-proliferative behaviour. Eur Radiol. 2019;29:1986–1996. doi: 10.1007/s00330-018-5704-8. [DOI] [PubMed] [Google Scholar]
- 17.Wang QY, Li QN, Mi R, et al. Radiomics nomogram building from multiparametric MRI topredict grade in patients with glioma: a cohort study. J Magn Reson Imaging. 2019;49:825–833. doi: 10.1002/jmri.26265. [DOI] [PubMed] [Google Scholar]
- 18.Trinchieri G. Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annu Rev Immunol. 2012;30:677–706. doi: 10.1146/annurev-immunol-020711-075008. [DOI] [PubMed] [Google Scholar]
- 19.Hasselbalch HC. Chronic inflammation as apromotor of mutagenesis in essential thrombocythemia, polycythemia vera and myelofibrosis. A human inflammation model for cancer development? Leuk Res. 2013;37:214–220. doi: 10.1016/j.leukres.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 20.Templeton AJ, McNamara MG, Šeruga B, et al. Prognostic role of neutrophil-to-lymphocyteratio in solid tumours: a systematic reviewand meta-analysis. J Natl Cancer Inst. 2014;23:1204–1212. doi: 10.1158/1055-9965.EPI-14-0146. [DOI] [PubMed] [Google Scholar]
- 21.Wan S, Lai Y, Myers RE, et al. Preoperative platelet count associates with survival and distantmetastasis in surgically resected colorectalcancer patients. J Gastrointest Cancer. 2013;44:293–304. doi: 10.1007/s12029-013-9491-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salvagno GL, Sanchis-Gomar F, Picanza A, et al. Red blood cell distribution width: A simple parameter with multiple clinical applications. Crit Rev Clin Lab Sci. 2015;52:86–105. doi: 10.3109/10408363.2014.992064. [DOI] [PubMed] [Google Scholar]
- 23.Bambury RM, Teo MY, Power DG, et al. The association of pre-treatment neutrophil to lymphocyte ratio with overall survival in patients with glioblastoma multiforme. J Neurooncol. 2013;114:149–154. doi: 10.1007/s11060-013-1164-9. [DOI] [PubMed] [Google Scholar]
- 24.Brockmann MA, Giese A, Mueller K, et al. Preoperative thrombocytosis predicts poor survivalin patients with glioblastoma. Neuro Oncol. 2007;9:335–342. doi: 10.1215/15228517-2007-013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auezova R, Ryskeldiev N, Doskaliyev A, et al. Association of preoperative levels of selectedblood inflammatory markers with prognosis in gliomas. Onco Targets Therapy. 2016;9:6111–6117. doi: 10.2147/OTT.S113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai J, Zhang C, Zhang W, et al. ATRX, IDH1-R132H and Ki-67 immunohistochemistry asa classification scheme for astrocytic tumors. Oncoscience. 2016;3:258–265. doi: 10.18632/oncoscience.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radiomics with artificial intelligence: a practical guide for beginners. Diagn Interv Radiol. 2019;25:485–495. doi: 10.5152/dir.2019.19321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim SM, Kim Y, Jeong K, et al. Logistic LASSO regression for the diagnosis of breast cancer using clinical demographic data and the BI-RADS lexicon for ultrasonography. Ultrasonography. 2018;37:36–42. doi: 10.14366/usg.16045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vasquez MM, Hu C, Roe DJ, et al. Least absolute shrinkage and selection operator typemethods for the identification of serum biomarkers of overweight and obesity: simulation and application. BMC Med Res Methodol. 2016;16:154. doi: 10.1186/s12874-016-0254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee EJ, Lee SK, Agid R, et al. Preoperative grading of presumptive low-grade astrocytomason MR imaging: diagnostic value of minimumapparent diffusion coefficient. AJNR Am J Neuroradiol. 2008;29:1872–1877. doi: 10.3174/ajnr.A1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ren Y, Pang HP, Feng XY, et al. Non-Gaussian diffusion MR imaging of glioma: comparisons of multiple diffusion parameters and correlation with histologic grade and MIB-1 (Ki-67 labeling) index. Neuroradiology. 2016;58:121–132. doi: 10.1007/s00234-015-1606-5. [DOI] [PubMed] [Google Scholar]
- 32.Smitha KA, Gupta AK, Jayasree RS. Total magnitude of diffusion tensor imaging as an effective tool for the differentiation of glioma. Eur J Radiol. 2013;82:857–861. doi: 10.1016/j.ejrad.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 33.Van Cauter S, Veraart J, Sijbers J, et al. Gliomas: diffusion kurtosis MR imaging in grading. Radiology. 2012;263:492–501. doi: 10.1148/radiol.12110927. [DOI] [PubMed] [Google Scholar]
- 34.Jiang R, Jiang J, Zhao L, et al. Diffusion kurtosis imaging can efficiently assess the glioma grade and cellular proliferation. Oncotarget. 2015;6:42380–42393. doi: 10.18632/oncotarget.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao J, Wang YL, Li XB, et al. Comparative analysis of the diffusion kurtosis imaging and diffusion tensor imaging in grading gliomas, predicting tumour cell proliferation and IDH-1 gene mutation status. J Neurooncol. 2019;141:195–203. doi: 10.1007/s11060-018-03025-7. [DOI] [PubMed] [Google Scholar]
- 36.Li FY, Shi WQ, Wang DW, et al. Evaluation of histopathological changes in the microstructure at the center and periphery of glioma tumors using diffusional kurtosis imaging. Clin Neurol Neurosurg. 2016;151:120–127. doi: 10.1016/j.clineuro.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 37.Jain KK, Sahoo P, Tyagi R, et al. Prospective glioma grading using single-dose dynamic contrast-enhanced perfusion MRI. Clin Radiol. 2015;70:1128–1135. doi: 10.1016/j.crad.2015.06.076. [DOI] [PubMed] [Google Scholar]
- 38.Jiang SJ, Hua Y, Zhou XJ, et al. Quantitative assessment of tumor cell proliferation in brain gliomas with dynamic contrast-enhanced MRI. Acad Radio. 2019;26:1215–1221. doi: 10.1016/j.acra.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 39.Tian Q, Yan LF, Zhang X, et al. Radiomics strategy for glioma grading using texture features from multiparametric MRI. J Magn Reson Imaging. 2018;48:1518–1528. doi: 10.1002/jmri.26010. [DOI] [PubMed] [Google Scholar]
- 40.Li YM, Qian ZH, Xu KB, et al. Radiomic features predict Ki-67 expression level and survival in lower grade gliomas. J Neurooncol. 2017;135:317–324. doi: 10.1007/s11060-017-2576-8. [DOI] [PubMed] [Google Scholar]
- 41.Weng YL, Zhang XY, Han JJ, et al. Do selected blood inflammatory markers combined with radiological features predict proliferation index in glioma patients? World Neurosurg. 2018;118:e137–146. doi: 10.1016/j.wneu.2018.06.142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1.
Characteristics of LGGs and HGGs patients in the training and validation cohorts
| Training cohort (n=107) | Validation cohort (n=45) | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| LGGs | HGGs | p | LGGs | HGGs | p | |
| Number | 36 | 71 | - | 11 | 34 | - |
|
| ||||||
| Age | ||||||
| Mean±SD | 44.53±11.31 | 51.93±22.68 | 0.001 | 40.18±8.85 | 56.79±12.27 | <0.001 |
|
| ||||||
| NLR | ||||||
| Median (Q1, Q3) | 2.07 (1.72, 3.21) | 2.73 (1.97, 4.64) | 0.036 | 1.83 (1.47, 2.89) | 3.21 (2.28, 5.12) | 0.016 |
|
| ||||||
| RDW | ||||||
| Mean±SD | 13.1±1.04 | 13.36±1.34 | 0.161 | 13.21±1.38 | 13.24±0.82 | 0.283 |
p < 0.05.
NLR, neutrophil-lymphocyte ratio; RDW, red cell distribution width; LGGs, low-grade gliomas; HGGs, high-grade gliomas.
Supplementary Table 2.
Characteristics of Grade III and IV patients in the training and validation cohorts
| Training cohort (n=74) | Validation cohort (n=31) | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Grade III | Grade IV | p | Grade III | Grade IV | p | |
| Number | 28 | 46 | - | 11 | 20 | |
|
| ||||||
| Age | ||||||
| Mean±SD | 50.18±13.30 | 54.72±12.44 | 0.099 | 51.27±13.52 | 26.6±13.25 | 0.082 |
|
| ||||||
| NLR | ||||||
| Median (Q1, Q3) | 3.04 (2.18, 4.63) | 2.8 (2.07, 4.72) | 0.608 | 2.25 (2.18, 2.6) | 3.20 (2.35, 6.42) | 0.043 |
|
| ||||||
| RDW | ||||||
| Mean±SD | 13.18±1.05 | 13.26±0.92 | 0.448 | 13.28±0.71 | 13.69±1.94 | 0.885 |
p < 0.05.
NLR, neutrophil-lymphocyte ratio; RDW, red cell distribution width.
Supplementary Table 3.
Characteristics of low Ki-67 and high Ki-67 patients in the training and validation cohorts
| Training cohort (n=107) | Validation cohort (n=45) | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Low Ki-67 LI | High Ki-67 LI | p | Low Ki-67 LI | High Ki-67 LI | p | |
| Number | 42 | 65 | - | 14 | 31 | - |
|
| ||||||
| Age | ||||||
| Mean±SD | 46.4±12.42 | 51.6±12.48 | 0.018 | 42.86±9.65 | 57.19±12.73 | 0.001 |
|
| ||||||
| NLR | ||||||
| Median (Q1, Q3) | 2.22 (1.75, 3.48) | 2.51 (2.07, 4.53) | 0.127 | 2.2 (2.34, 5.4) | 3.42 (2.34, 5.4) | 0.013 |
|
| ||||||
| RDW | ||||||
| Mean±SD | 13.01±0.84 | 13.44±13.0 | 0.116 | 13.17±1.24 | 13.26±0.84 | 0.233 |
p < 0.05.
NLR, neutrophil-lymphocyte ratio; RDW, red cell distribution width; Ki-67 LI, Ki-67 labeling index.
Compared with the individual sequences, the ROC curves showed the effectiveness of the multiparametric MRI in differentiating between LGGs and HGGs, grade III and grade IV gliomas, and low Ki-67 and high Ki-67 in the training cohort (a–c) and validation cohort (d–f), respectively.