Figure 8.
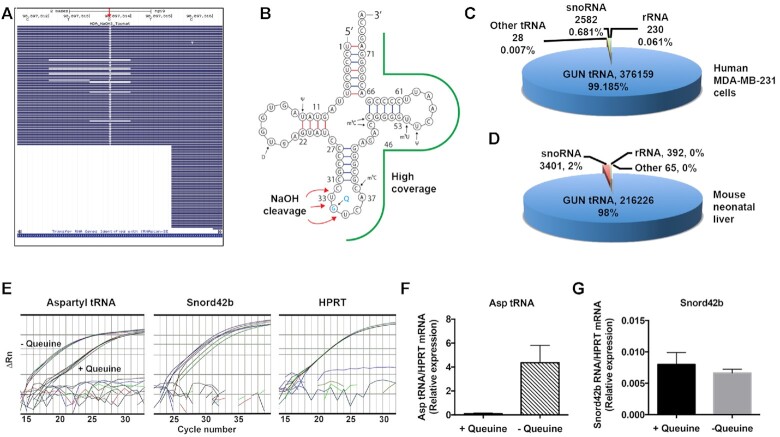
GUN tRNA are the principle small RNA substrates of the QTRT enzyme. RNA from human MDA-MB-231 cells and neo-natal mouse liver was processed by the capture and release method, adapter ligated and subjected to NGS sequencing on a MiSeq platform. Human and mouse fastq files were aligned using Tophat2 to the hg19 and mm10 genome assemblies, respectively. (A) Representative BAM file for an MDA-MB-231 alignment, visualized on the UCSC genome browser, showing the canonical ‘TGTN’ site—UGUC in aspartyl tRNA—showing the G to T transversion (red arrow) created by the abasic site during library generation. (B) Aspartyl tRNA sequence showing the regions of high coverage (green outline) and the sites of NaOH cleavage (red arrows). (C andD) Representative pie-charts showing the read number and percentage of RNA species captured from MDA-MB-231 cells and neo-natal mouse liver from a single run. For example, assay to determine if snoRNA can serve as in vivo substrate for the QTRT enzyme. Splenocytes recovered from 8-week-old, EAE-disease induced C57BL/6J female mice upon reaching a clinical score of 2. Single cell suspensions (2 × 106 cells/ml) in X-vivo 15 medium were re-stimulated with MOG[33–55] (50 μg/ml) for 48 h. Cells were left untreated or administered a large excess of queuine (200 μM) for 24 h before total RNA was isolated. (E) Following the capture-release method, Aspartyl tRNA and snord42b RNA quantified by stem-loop adapter-ligated RTPCR and adapter-ligated Taqman RTPCR, respectively. HPRT mRNA quantified by Taqman RTPCR directly from total RNA. (F andG) Ratio of aspartyl tRNA and Snord42b RNA relative to HRPT mRNA transcript levels.