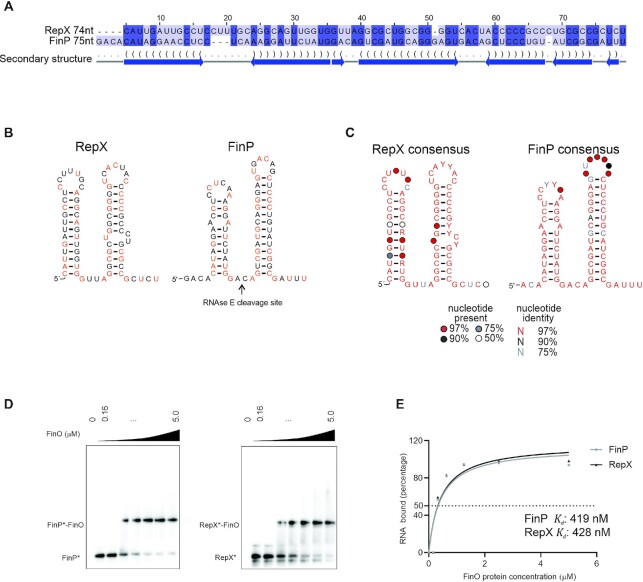
Figure 4.
FinO specificity for FinP-like structured sRNAs. (A) FinP and RepX sequences from pSLT and pRSF1010 were aligned in LocARNA (44) and visualized on Jalview (47). (B) Predicted structure of FinP and RepX sRNAs. In orange indicated nucleotides that are conserved between FinP and RepX. (C) RepX and FinP consensus structure from alignment in Supplementary Figure S2. (D) EMSA with FinO of FinP and RepX sRNAs. Radiolabeled FinP (left panel) or RepX (right panel) (4 nM) was incubated with increasing concentration of FinO (0, 0.16, 0.32, 0.63, 1.25, 2.5, 5 μM). (E) Quantification of FinO binding affinity to FinP and RepX. The apparent Kd of FinO binding to FinP or RepX was determined by quantifying the intensity of the bound and unbound fraction by FinO for FinP and RepX on ImageJ. The percentage of RNA bound is plotted as a function of FinO concentration. The values were fitted to a curve followed by a nonlinear regression. The apparent Kd was determined as the concentration at which 50% of the labeled FinP* or RepX* is bound by FinO in our assay.