Table 2.
Ablation studies on GraphBind with different settingsa
| Posb | PEc | rgd | rve | EUf | Ag | GRUh | NLi | Dj | Rec | Pre | F1 | MCC | AUC | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Base | C | T | 20 | 10 | T | S | T | 4 | 128 | 0.676 | 0.537 | 0.598 | 0.558 | 0.926 |
| (A) | SC | 0.593 | 0.591 | 0.592 | 0.552 | 0.922 | ||||||||
| CA | 0.633 | 0.537 | 0.581 | 0.538 | 0.921 | |||||||||
| (B) | F | 0.650 | 0.528 | 0.583 | 0.540 | 0.920 | ||||||||
| (C) | 15 | 0.634 | 0.551 | 0.589 | 0.548 | 0.919 | ||||||||
| 25 | 0.656 | 0.540 | 0.593 | 0.551 | 0.923 | |||||||||
| 30 | 0.580 | 0.594 | 0.587 | 0.547 | 0.913 | |||||||||
| (D) | 5 | 0.622 | 0.472 | 0.537 | 0.490 | 0.910 | ||||||||
| 13 | 0.663 | 0.540 | 0.595 | 0.555 | 0.923 | |||||||||
| (E) | F | 0.570 | 0.483 | 0.523 | 0.474 | 0.899 | ||||||||
| (F) | M | 0.561 | 0.407 | 0.472 | 0.418 | 0.875 | ||||||||
| (G) | 2 | 0.630 | 0.524 | 0.573 | 0.529 | 0.914 | ||||||||
| 3 | 0.647 | 0.551 | 0.595 | 0.554 | 0.925 | |||||||||
| 5 | 0.647 | 0.545 | 0.592 | 0.550 | 0.925 | |||||||||
| 6 | 0.688 | 0.522 | 0.586 | 0.545 | 0.924 | |||||||||
| F | 2 | 0.670 | 0.523 | 0.587 | 0.546 | 0.925 | ||||||||
| F | 4 | 0.637 | 0.541 | 0.585 | 0.543 | 0.922 | ||||||||
| F | 6 | 0.669 | 0.504 | 0.575 | 0.533 | 0.922 | ||||||||
| (H) | 64 | 0.593 | 0.527 | 0.558 | 0.513 | 0.910 |
aOnly different settings are given and other settings (empty values) are the same as the base model. These metrics are calculated on the validation set of DNA-573_Train and the highest values are bolded.
bPseudo-position of a residue: C, SC and CA stand for the centroid of residue, the centroid of residue side-chain and the position of alpha-C atom, respectively.
cUse the relative distance from every node to the sphere center as position embeddings of nodes (T) or not (F).
dRadius of the structural context: it defines the nodes belonging to a graph of a residue, and its unit is Å.
eThe threshold of adjacent matrix: it binarizes a distance matrix to the adjacent matrix to define the adjacent edges belonging to a node, and its unit is Å.
fUse the edge feature vectors (T) or not (F).
gThe aggregation operation in the node update module and the graph update module. S and M stand for sum and max operation, respectively.
hUse GRU (T) or not (F). If GRU is not used, the output 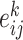 ,
, ,
,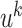 equal the intermediate output
equal the intermediate output  ,
, 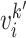 ,
, 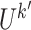 , respectively.
, respectively.
iThe number of GNN-blocks.
j
D
e, Dv and Du stand for the dimension of encoded edge feature vectors, the dimension of encoded node feature vectors and the dimension of encoded graph feature vectors, respectively. We set De Dv
Dv Du.
Du.
