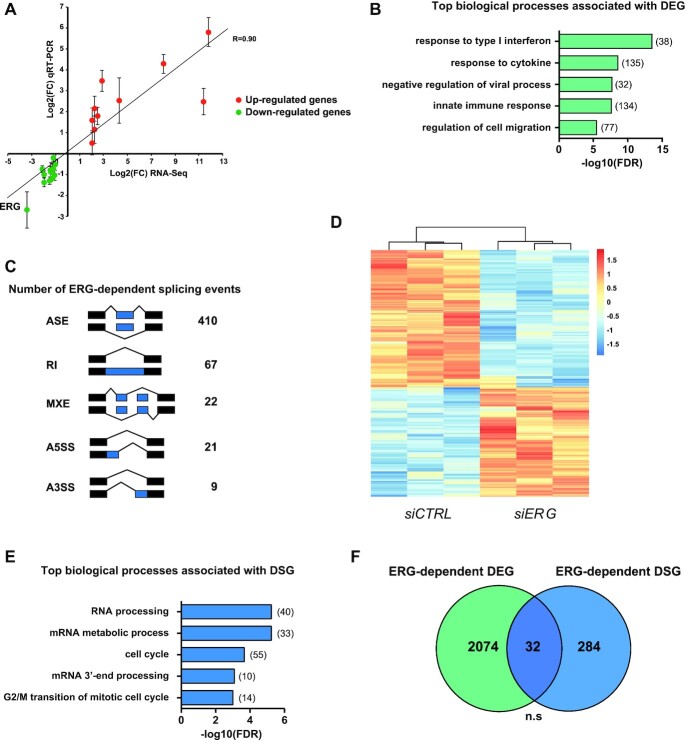
Figure 1.
ERG controls the HeLa transcriptome at the levels of gene expression and alternative splicing. (A) Correlation between mRNA levels quantified by RNA-sequencing or qRT-PCR on a set of 22 target mRNAs expected to be up- (FDR<0.05, Log2(FC)>1, red dots) or down- (FDR < 0.05, log2(FC) < 1, green dots) regulated after ERG knockdown in HeLa cells. Results shown are means ± SD (n = 3 independent experiments). (B) Distribution of enriched GO biological process terms in differentially expressed genes (DEG) following ERG knockdown in HeLa cells. (C) Numbers of significantly differentially spliced events identified by rMATS after ERG knockdown in HeLa cells with the following criteria: supported by at least 15 unique reads, |ΔPSI| > 10% and FDR < 0.05. ASE: Alternatively spliced exons, RI: Retained intron, MXE: Mutually exclusive exons, A5SS: Alternative 5′ splice site, A3SS: Alternative 3′ splice site. (D) Heatmap of Z-scores of percent of spliced-in (PSI) values from significantly differentially spliced exons between HeLa cells transfected either with siCTRL or siERG. (E) Distribution of enriched GO biological process terms associated with differentially spliced genes (DSG, i.e. genes with at least one ERG-regulated ASE) following ERG knockdown in Hela cells. (F) Overlap between differentially expressed genes and differentially spliced genes (DSG) in HeLa cells after ERG knockdown. Expected overlap = 39. n.s.: not significant.