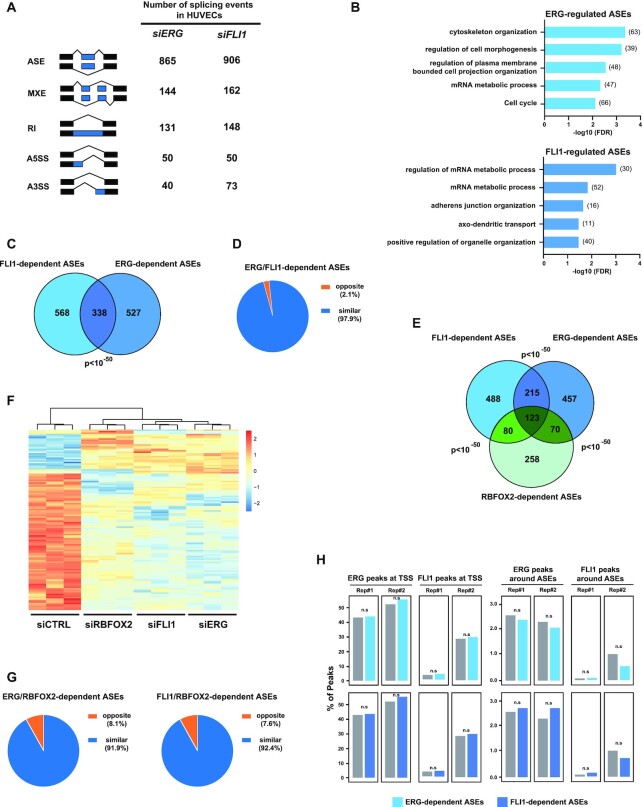
Figure 4.
ERG and FLI1 shape the transcriptome at the level of gene expression and mRNA splicing in HUVECs. (A) Numbers of significantly differentially spliced events identified after ERG or FLI1 knockdown in HUVECs using rMATS with the following criteria: supported by at least 15 unique reads, |ΔPSI| > 10% and FDR < 0.05. ASE: Alternatively spliced exons, RI: Retained intron, MXE: Mutually exclusive exons, A5SS: Alternative 5′ splice site, A3SS: Alternative 3′ splice site. (B) Distribution of enriched GO biological process terms associated with differentially spliced genes (DSG, i.e. genes with at least one ERG-regulated ASE) following ERG (top) or FLI1 (bottom) knockdown in HUVECs. (C) Overlap of significantly differentially spliced exons modulated upon ERG or FLI1 knockdown in HUVECs. Expected overlap = 14. (D) Proportion of common target exons upon ERG or FLI1 inhibition shown in (C) (n = 338) categorized as ‘similar’ (blue sector) or ‘opposite’ (orange sector) depending on whether delta PSI values vary in respectively the same or opposite direction. (E) Overlap of significantly differentially spliced exons modulated upon ERG, FLI1 or RBFOX2 knockdown in HUVECs. Expected overlap: FLI1/ERG = 14, FLI1/RBFOX2 = 8, ERG/RBFOX2 = 8. (F) Heatmap of Z-scores of percent of spliced-in (PSI) values from common significantly differentially spliced exons between HUVEC cells transfected either with siERG, siFLI1 or siRBFOX2, relative to siCTRL. (G) Proportion of common target exons upon ERG and RBFOX2 or FLI1 and RBFOX2 inhibition in HUVECs shown in (E) (n = 193 and n = 203, respectively) categorized as ‘similar’ (blue sector) or ‘opposite’ (orange sector) depending on whether delta PSI values vary in respectively the same or opposite direction. (H) We compared genes with no ASEs (control group) and genes with ASEs (ERG-dependent in light blue and FLI1-dependent in dark blue). The barplots represent the proportion of genes that have a ERG or FLI1 ChIP peak on either TSS (left panel) or around exons (±250 bp, right panel). n.s: not significant, by chi-square test.