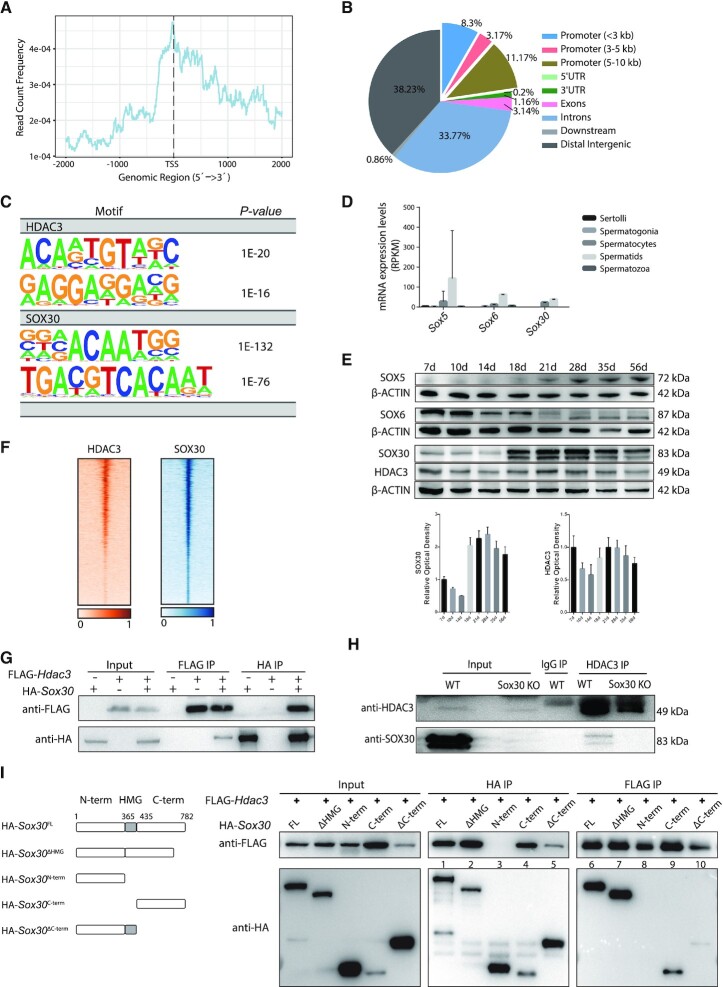
Figure 6.
HDAC3 and SOX30 co-localize on the genome in testes. (A) Normalized HDAC3 ChIP-seq reads on UCSC mm10 RefSeq gene bodies. (B) Annotations of HDAC3 ChIP-seq peaks in wild-type testes at P20. (C) Top enriched motifs in the binding sites for HDAC3 and SOX30 using Homer. Sequences within ± 200 bp from the centers of all the binding sites were used for de novo motif analysis. (D) mRNA transcripts levels of Sox members across stage-specific spermatogenic cells were assessed by RNA-seq (14). (E) Western blot analysis for the protein levels of SOX5, SOX6, SOX30, and HDAC3 during spermatogenesis. The image is a representation of two independent experiments with similar results. The corresponding optical density readings for biological duplicates are shown. (F) Heat map depicting HDAC3 and SOX30 co-localization at many binding sites. (G) FLAG-Hdac3 and/or HA-Sox30 expression constructs were transfected in HEK 293T cells and subjected to immunoprecipitation analysis. (H) Testes protein lysates were immunoprecipitated either with HDAC3 or normal IgG antibodies followed by immunoblot analysis. Protein lysates were prepared from wild type and Sox30 KO at postnatal day 20. (I) HA-tagged SOX30 mutants with deleted fragments were coexpressed with FLAG-HDAC3 in HEK 293T cells. Lysates were immunoprecipitated with anti-FLAG antibodies, and the immunoprecipitated protein complex was examined for HA (SOX30).