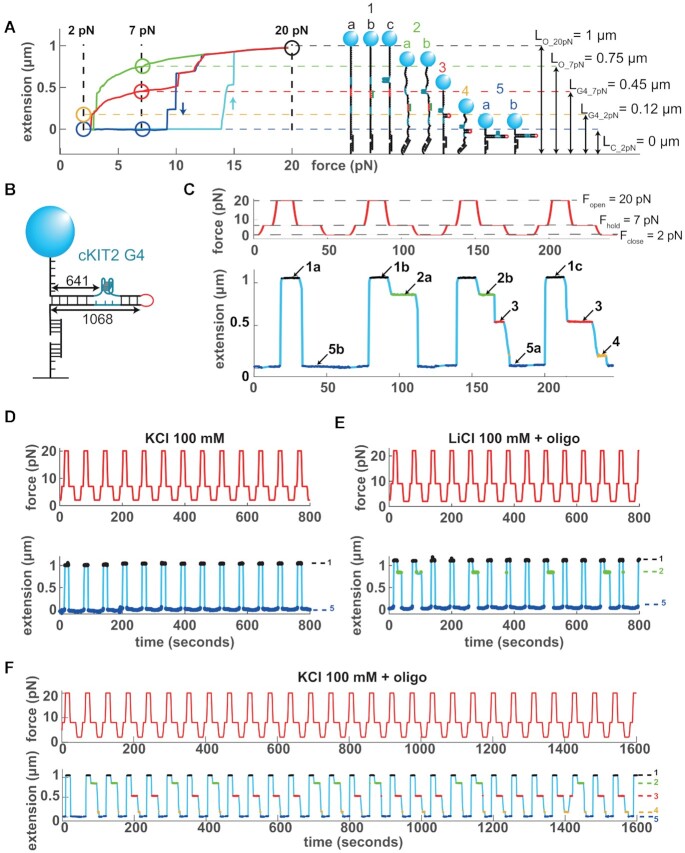
Figure 1.
Single molecule assay for detection of G4 formation. (A) Left panel: Force-extension curves of a 1068 bp DNA hairpin containing a cKIT-2 G4 forming sequence. Open circles mark the different molecule extensions observed at the three test forces. Cyan: hairpin opening at increasing forces. Blue: hairpin closing with no G4 or blocking oligonucleotide at decreasing forces. Green: hairpin closing with a blocking oligonucleotide. Red: hairpin closing with a folded G4. LO − 20 pN: fully open hairpin at 20 pN. LO − 7 pN: oligonucleotide-blocked hairpin at 7 pN. LG4 − 7 pN: G4-blocked hairpin at 7 pN. LG4 − 2 pN: G4-blocked hairpin at 2 pN. LC − 2 pN: fully closed hairpin at 2 pN. Right panel: structures inferred from the force-extension curves. (B) Schematics of the hairpin molecule, indicating the size in bp of the full dsDNA hairpin and the position of the G4 sequence. (C) Force cycles (red) and extension traces (blue) illustrating the corresponding molecular states described in (A). (D) Force cycles (800 s, red) and corresponding extension traces (blue) of cKIT-2 hairpin in KCl only (color-coded numbers correspond to structures described in (A)). (E) Same as (D) in 100 mM LiCl plus blocking oligonucleotide. (F) Same as (D) in 100 mM KCl with blocking oligonucleotide.