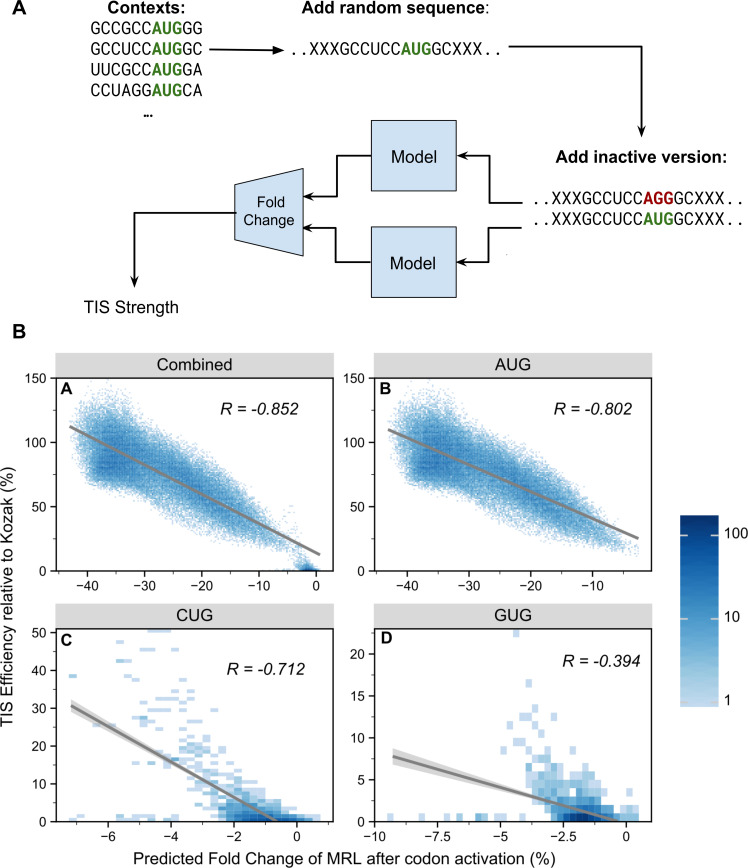
Fig 3. Framepool predicts the strength of upstream transcription initiation sites.
A) The model was used to predict the effect of upstream TIS contexts by injecting them into random sequences and comparing predicted MRL fold change associated with upstream start codon activation (mutation AGG → AUG). B) These predictions (x-axis) were correlated with independent experimental measurements of the relative strength of these contexts (y-axis). Overall, a negative correlation is observed (top right) as expected because upstream TISs compete with canonical TISs. For AUG start codon data [9], the model correlates well with the strength estimates (top right, Pearson correlation of -0.802), with more efficient contexts causing a larger predicted effect on MRL. Negative correlations were also observed for the alternative start codons CUG (bottom left) and GUG (bottom right) data [31]. The correlations are weaker, particularly for GUG, likely reflecting the difficulty of detecting more subtle motifs in MPRA data. Nevertheless, the model has still learned to distinguish between strong and weak CUG/GUG motifs to some extent.