Abstract
Polyploidy has been hypothesized to be both an evolutionary dead-end and a source for evolutionary innovation and species diversification. Although polyploid organisms, especially plants, abound, the apparent nonrandom long-term establishment of genome duplications suggests a link with environmental conditions. Whole-genome duplications seem to correlate with periods of extinction or global change, while polyploids often thrive in harsh or disturbed environments. Evidence is also accumulating that biotic interactions, for instance, with pathogens or mutualists, affect polyploids differently than nonpolyploids. Here, we review recent findings and insights on the effect of both abiotic and biotic stress on polyploids versus nonpolyploids and propose that stress response in general is an important and even determining factor in the establishment and success of polyploidy.
Introduction
Polyploidy, that is having multiple sets of chromosomes as a consequence of whole-genome duplication (WGD), has been studied for over 100 years, especially in flowering plants, dating back to the classic work of Hugo de Vries, Øjvind Winge, Cyril Darlington, and G. Ledyard Stebbins, Jr (Soltis et al., 2014a). Because of the well-known detrimental effects arising from doubling one’s entire chromosome set, most WGD events are not successful, as new polyploids are typically not better adapted than their diploid parent(s). Genomic instability, mitotic and meiotic abnormalities, and minority cytotype exclusion (Levin, 1975; Comai, 2005; Madlung et al., 2005; Morgan et al., 2020) are all expected to quickly remove new polyploids from the population. Nevertheless, there are numerous polyploid organisms around us; even those currently considered diploids usually bear signatures of a polyploid ancestry (Wendel, 2015; Van de Peer et al., 2017). Furthermore, several of these ancestral polyploidy events can be traced back to the origin and diversification of major plant lineages, including angiosperms, core eudicots, monocots, orchids, grasses, composites, and legumes (Van de Peer et al., 2017). The same holds true in animals, such as vertebrates, and fishes (Taylor et al., 2003; Le Comber and Smith, 2004; Dehal and Boore, 2005), suggesting an important role for polyploidy in promoting phenotypic diversity (Soltis and Soltis, 2009; Landis et al., 2018; Leebens-Mack et al., 2019). In fact, polyploidy is now generally considered to be a common mode of speciation with far-reaching ecological and evolutionary consequences, not only in plants, but also in many other eukaryotes. In addition, polyploid organisms or polyploid populations are often considered more resilient to extreme environments because of their increased genetic variation and the buffering effect of their duplicated genes, which has led to an increased recognition of the short-term adaptive potential of polyploidy (Van de Peer et al., 2009, 2017; Doyle and Coate, 2019).
The number of extant plant species recognized as polyploids exceeds that of ancient WGDs by several orders of magnitude (Van de Peer et al., 2017), although many additional ancient WGDs continue to be uncovered by applying genomic and transcriptomic deep sequencing methods to more lineages of life (Godden et al., 2019; Leebens-Mack et al., 2019). Nevertheless, because relatively recent (i.e. the last few million years) polyploid species are so prevalent, one might expect to find evidence of substantially more ancient WGDs. The many examples of recurrent polyploidy (Soltis and Soltis, 1999), the existence of many polyploids of fairly recent origin (Barker et al., 2016), and mounting evidence of widespread cryptic polyploid species (Soltis et al., 2007; Laport and Ng, 2017) would appear to contrast with the evidence of relatively few ancient polyploidy events, certainly within the same lineage (Van de Peer et al., 2017; Wong et al., 2020). This result is perhaps not surprising in that polyploidy is prevalent, but not all successful polyploidizations will ultimately lead to major species diversification over the longer term (Arrigo and Barker, 2012; Soltis et al., 2015). Notwithstanding, most plant lineages show evidence for one or few established ancient WGDs (Leebens-Mack et al., 2019; Wong et al., 2020). For example, all extant angiosperms have at least one ancient WGD in their ancestry, with some lineages having experienced several additional rounds of genome doubling over time. What is fascinating, however, is that the establishment or long-term survival of many of these WGDs is not random, but instead coincides with major periods of global climatic/geologic change and/or periods of mass extinction (Van de Peer et al., 2017; Novikova et al., 2018; Cai et al., 2019; Koenen et al., 2020; Wu et al., 2020), i.e. periods of major stress (Figure 1).
Figure 1.
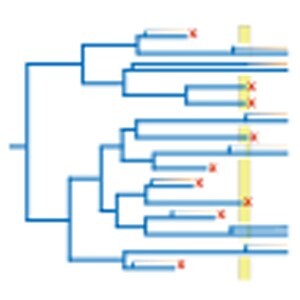
The nonrandom establishment of WGDs. Red crosses represent extinction, while orange circles represent successful polyploidy coinciding with environmental stress, and light orange triangles represent successful polyploidy following delayed rediploidization [responsible for lag times between the WGD event and its exerted effects {Schranz et al., 2012; Robertson et al., 2017}]. The dark blue diamond represents a diploid having survived environmentally challenging conditions. The two branches descending from a WGD event represent the duplicated genome, while the changing color in one branch denotes the divergence (and diploidization) of the subgenomes. Gray squares represent WGDs that coincide (purple) or seem to coincide (light gray) with a period of global change or extinction. If polyploidy by itself (without rediploidization, functional divergence of genes, or rewiring of networks) enables evolutionary innovation (e.g. through short-term gene expression changes, epigenetic remodeling), WGDs might become established even in the absence of rediploidization (represented by the light blue square). See text for details.
The recurring association involving polyploidy, gene duplication, and stress (Doyle and Coate, 2019) is evident at multiple levels and stages. From an agricultural standpoint, duplicated genes resulting from polyploidy appear to have been key to crop domestication and the evolution of stress resistance (Renny-Byfield and Wendel, 2014). It is also well-known that stress can trigger polyploidy (Ramsey and Schemske, 1998); concomitantly, there are also strong indications that polyploidy confers a selective advantage under stressful or changing environmental conditions (Van de Peer et al., 2017). Moreover, an increasing number of studies describes how biotic stress and interactions with, for instance, pathogens and herbivores, affect polyploids in different ways than their related diploids (Hannweg et al., 2016; Hias et al., 2018). In addition, cellular polyploidy (endoreduplication) is an induced response to stress in normally diploid organisms that may mitigate the effects of stress (Pacey et al., 2020a; Zedek et al., 2020).
In this Review, we discuss the connection between polyploidy (both auto- and allo-polyploidy) and stress, both in terms of formation and survival. We emphasize current views on how polyploidy might confer an advantage when dealing with both abiotic and biotic stress. By stress, we mean strain on an organism resulting from adverse or demanding circumstances and more specifically environmental factors that cause stress, including abiotic factors (e.g. soil, temperature, water availability) and biotic factors (e.g. predators, pathogens). We highlight recent studies that provide a better understanding into how and why polyploids might better adapt to changing environments, why they might be able to escape periods of extinction better than diploids, and why they respond to biotic agents differently than diploids. We will not discuss—at least not in any detail—the many different ways in which gene or genome duplicates can evolve novel gene functions (neofunctionalization) or acquire tissue-specific or temporally segregated functions (subfunctionalization), how remnants of ancient duplication events can be discovered, and how a polyploid genome undergoes fractionation and diploidizes. We will instead refer the reader to several excellent publications on these topics (e.g. Wolfe, 2001; Taylor and Raes, 2004; Van de Peer, 2004; Conant and Wolfe, 2008; Blischak et al., 2018; Mandakova and Lysak, 2018; Wendel et al., 2018; Doyle and Coate, 2019).
Polyploidy and the stress connection
Polyploidy and abiotic stress
Signs from the past
WGD is often considered an evolutionary dead-end, particularly under stable conditions (Comai, 2005; Oberlander et al., 2016; Van de Peer et al., 2017; Figure 1). However, ancient WGDs are not randomly distributed over time, but rather appear to overlap with periods of dramatic global change, the most apparent of which coincides with the Cretaceous-Paleogene boundary (K-Pg; Fawcett et al., 2009; Vanneste et al., 2014; Cannon et al., 2015; Levin and Soltis, 2018; Koenen et al., 2020). Several possible explanations come to mind. First, the opportunity for the formation of new polyploid species might increase subsequent to a cataclysmic event. It has long been recognized that the levels of unreduced gametes can increase in response to external stimuli, such as environmental stress (Bretagnolle and Thompson, 1995; Ramsey and Schemske, 1998). A high frequency of unreduced gametes is associated with temperature stress in fish and amphibians (Mable et al., 2011), and particularly in plants (Ramsey and Schemske, 1998; Mason et al., 2011; Pecrix et al., 2011; De Storme et al., 2012). The effect of stress on unreduced gamete formation is not limited to present-day plants. Indeed, increased levels of unreduced pollen from fossil plants were observed at the Triassic–Jurassic transition, which corresponds to the fourth of the five major extinction events (Kurschner et al., 2013), while abnormal gymnosperm pollen (Foster and Afonin, 2005) and lycophyte spores (Visscher et al., 2004) have been reported during the Permian–Triassic transition, corresponding to the third major extinction event. Second, during stressful times or during times of mass extinction, WGDs have an elevated probability of successful establishment because of increased mutational robustness as well as increased genetic and epigenetic variation, in turn raising the adaptive potential of a polyploid species. The buffering effect against (deleterious) mutations, altered gene expression, epigenetic remodeling, sub-, and/or neofunctionalization, and/or the rewiring of more complex gene regulatory networks (GRNs) following WGD can subsequently provide a selective advantage to the new polyploid in a changed environment, possibly through the derivation of novel traits. In both cases (the origin of new polyploids and the persistence of polyploid lineages), the dating of WGD events will coincide with the period of environmental change (Figure 1). However, for autopolyploids, Robertson and co-authors suggested that delayed diploidization might explain “lag-times” between the occurrence of WGDs and their potential effects (Robertson et al., 2017). Delayed diploidization occurs as long as any two of the four chromosome sets resulting from a WGD pair randomly during meiosis, leading to recombination between them and thus preventing sequence and functional divergence between homoeologs (also called ohnologs). Consequently, homoeolog divergence as well as the potential for evolutionary innovation is also delayed (Figure 1). Only during or after chromosomal diploidization (leading to disomic inheritance) can the subgenomes start to diverge (as do their genes), causing a putative evolutionary effect or selective advantage of WGD. Therefore, the “Lineage-specific Ohnolog Resolution” or “LORe” model may provide a plausible framework to explain how the functional and evolutionary outcomes of genome doubling, and particularly its contribution to extinction avoidance, may be constrained from arising for millions or tens of millions of years after the WGD event (Schranz et al., 2012; Robertson et al., 2017). Thus, WGDs with delayed diploidization, as predicted by the LORe model, may be predicted to be quite a bit younger than they actually are, namely at the age they exert an effect—e.g. during environmental turmoil—rather than at the time they occurred. Therefore, the so-called “waves” of ancient polyploidy, coinciding with challenging or stressful conditions, might also be artificially enriched with WGDs (Figure 1).
Several other, more recent epoch boundaries have also been proposed to align with periods of extensive WGD (Estep et al., 2014; Kagale et al., 2014; Huang et al., 2016; Novikova et al., 2018; Cai et al., 2019; Sessa, 2019). Analysis of WGD events across seed plants (Leebens-Mack et al., 2019) indicates multiple waves of genome doubling corresponding not only to the K-Pg, but also the Middle Miocene Disruption as well as the Paleocene–Eocene Thermal Maximum (Soltis and Landis, unpublished data). The clustering of polyploidy events in association with worldwide, geologically significant events seems to affirm the importance of environmental conditions in the promotion/survival of polyploidy (Levin, 1983; Soltis and Soltis, 2014; Doyle and Coate, 2019).
Signs from the present
The close relationship between stress and polyploidy is also supported by present-day polyploids; there is a long history of association between polyploidization and abiotic stress, particularly adaptation to extremely dry and cold environments (Stebbins, 1949; Ehrendorfer, 1980; Levin, 1983; Folk and Soltis, 2020; Lourkisti et al., 2020). Numerous studies have suggested an association between dry habitats and polyploidy across diverse lineages of land plants, with polyploids occupying drier habitats than observed for the diploids in those same clades (te Beest et al., 2012; Gunn et al., 2020). Polyploidy increases in frequency from the equator to the poles (Rice et al., 2019); within the Arctic, the frequency of higher order polyploids increases with latitude (Brochmann et al., 2004). In the context of adaptation to drought stress, genome doubling has been shown to lead to changes in transpiration, water use efficiency, photosynthetic rate, phenology, antioxidant response, and morphology (Maherali et al., 2009; Deng et al., 2012; Soltis and Soltis, 2014). For example, naturally occurring tetraploid Arabidopsis (Arabidopsis thaliana) plants exhibit increased salt tolerance compared to diploids (Chao et al., 2013). Both tetraploid rice (Oryza sativa) and citrange (Citrus sinensis L. Osb. x Poncirus trifoliata L. Raf.) have an increased tolerance to salt and drought stress as a result of WGD, which affects the expression of genes involved in stress and phytohormone response pathways (Yang et al., 2014b; Ruiz et al., 2016). Similarly, tetraploid rootstock-grafted watermelon (Citrullus lanatus) plants are more tolerant to salt stress than are diploid plants (Zhu et al., 2018). Although the physiological, cellular, and genomic responses to stress are more frequently documented for polyploids (see further), the exact molecular processes underlying these responses still remain to be discovered (Fox et al., 2020).
Polyploidy and Stress, a Universal Connection?
Higher abiotic stress tolerance has also been demonstrated in fungi and animals. For instance, experimental evolution studies in yeast (Saccharomyces cerevisiae) have shown that genome duplication can confer a fitness advantage during stressful conditions (Selmecki et al., 2015). Diploid frog species of the genus Neobatrachus are currently geographically isolated, whereas tetraploid Neobatrachus species can occupy harsher environments and are distributed more widely across Australia; polyploidy-mediated gene flow and hybridization may promote the adaptive advantage of the tetraploids in the face of climate change (Novikova et al., 2020). Another example involves the initial establishment and subsequent radiation of polyploidy in cyprinid fish, likely triggered by environmental and climate changes associated with the uplift of the Tibetan Plateau. Moreover, uniquely retained homoeologous genes that confer adaptation to harsh environments were identified (Li and Guo, 2020). WGD in an ancestor of the giant African snail (Achatina immaculata) was estimated to have occurred ∼70 Mya, possibly coinciding with the K-Pg extinction event (Liu et al., 2020). Although terrestrial adaptation of these snails was initiated before WGD, specific homoeologs encoding hemocyanins, involved in O2 transport, and genes involved in gluconeogenesis were retained after WGD, enhancing the capacity for gas exchange and glucose homeostasis in aestivation (animal dormancy during a dry or hot season). WGD may have facilitated terrestrial adaptation by increasing the survival rate in the stressful transition from water to land (Liu et al., 2020).
Polyploidy and biotic stress
Plant–antagonist interactions
Aside from abiotic stress, organisms experience biotic stress in various forms, including attacks by pathogens, predators, or herbivores, and competition with other species of the same trophic level. Changes in anatomical structures, growth regulators, and other physiological processes associated with WGD are predicted to alter these interactions and may increase the ability of a polyploid to resist or tolerate damage, or to garner shared resources. For instance, models predict that polyploidy can increase resistance within the gene-for-gene systems that underlie many host–pathogen interactions or where genotype × genotype interactions are important (Oswald and Nuismer, 2007). In a test of this model, synthetic neopolyploids of a monogenic resistant apple cultivar (Malus × domestica) exhibited increased resistance to apple scab (Venturia inaequalis) compared to diploid cultivars (Hias et al., 2018). Another study found that synthetic tetraploid garden impatiens (Impatiens walleriana) showed improved resistance to downy mildew (Plasmopara obducens) relative to diploid plants (Wang et al., 2018b). Similarly, Hannweg et al. found that synthetic tetraploids of the Livingstone potato (Plectranthus esculentus) were more resistant to root-knot nematodes than diploids (Hannweg et al., 2016). Comparable findings have been reported in animal systems. In experimental (bath) inoculations of diploid and triploid Atlantic salmon (Salmo salar L.) with salmonid alphavirus, symptoms of the disease appeared more slowly in the triploids than in the diploids, although there was no difference in viral loads between ploidies (Moore et al., 2017). Interestingly, Harms et al. (2020) found in lab bioassays that triploid genotypes of the Eurasian wetland weed, Butomus umbellatus, had larger leaf lesions than diploids in response to several pathogens (Keane et al., 2014), indicating their lower resistance compared to diploid genotypes, although in the field triploids had ∼50% lower disease incidence than diploids, suggesting that plant–pathogen interactions may be modified by climatic differences in pathogenicity or in the regional pool of pathogens. Pathogens themselves may also shift ploidy during infections, such as with Phytophthora infestans that led to the Great Irish potato famine (Li et al., 2017) or, more recently, a pathogen that increasingly infects European honeybees (Peters et al., 2019), evoking the possibility for ploidy-driven coevolutionary dynamics.
Increased abiotic tolerance exhibited by polyploids may make them better competitors than diploid relatives, especially under limited resources or marginal habitats (Segraves, 2017; Baduel et al., 2018; Harms et al., 2020). For instance, while triploid genotypes of the Eurasian wetland weed, Butomus umbellatus, were more poorly adapted than diploids under some conditions, Harms et al. concluded that their increased root: shoot ratios made them better-adapted competitors than diploids over several years (Harms et al., 2020). In a 5-year study in China with diploid, tetraploid, and hexaploid canadian goldenrod (Solidago canadensis), polyploidization and rapid evolution post-introduction contributed to increased competitive ability in polyploids and influenced the outcome of community succession (Wu et al., 2019).
Biotic interactions can ameliorate stress
How plants respond to abiotic stresses such as drought or limited essential nutrients, or to biotic stresses such as pathogens and herbivores (Figure 2), may be mediated by mutualistic interactions with bacteria (e.g. rhizobia that inhabit root nodules) and fungi (e.g. root-associating mycorrhizae or leaf endophytes; for review, see Acuna-Rodriguez et al., 2020). If polyploids have enhanced interactions with mutualists, they may then be able to tolerate stresses that their related diploids cannot. In this way, mutualisms may also contribute to polyploid success by enabling or reinforcing abiotic niche differentiation from diploids. For instance, theory shows that increasing mutualistic partner breadth can expand the range of abiotic tolerances for a species (Batstone et al., 2018) and thus may facilitate the ability of a polyploid to differentiate its abiotic niche from that of diploids (a potential prerequisite for polyploid speciation (Thompson and Lumaret, 1992). Moreover, if polyploids modify the local soil microbiome in ways that feed back to enhance their own growth, but not that of diploids, they may amplify their own persistence while suppressing other species, as predicted by niche construction theory (Albuquerque et al., 2019).
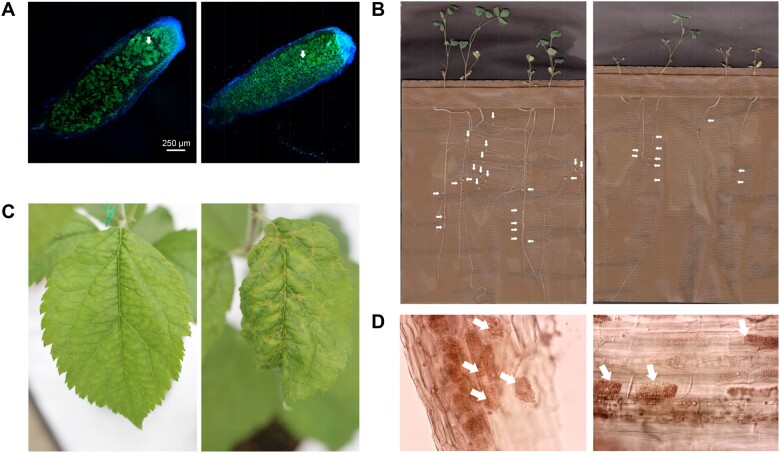
Figure 2.
Representative examples of the effects of polyploidy on mutualistic and antagonistic species interactions. (A, B, and D), Examples of mutualistic interactions. (C), Antagonist interaction. (A) Root nodules of synthetic neotetraploid (left) and diploid (right) alfalfa (Medicago sativa subsp. caerulea) inoculated with rhizobia (Forrester and Ashman, 2020). The autotetraploid plants produced nodules that were 20% larger than diploids and these housed symbiosomes—each with enlarged bacteroids (white arrows)—that were nearly twice the size of those present in diploids. Nodules are stained with SYTO 13 bacteroid fluorescence is depicted in green and plant autofluorescence in blue, 10× lens was used for viewing. (B) Tetraploid (left) alfalfa subsp. sativa and diploid (right) alfalfa subsp. caerulea showing root nodule production (Forrester et al., 2020). Autotetraploid alfalfa produced more than twice as much total root nodule biomass (white arrows pointing to individual nodules) than diploids, and this leads to higher shoot biomass. (C) Tetraploid (left) and diploid (right) apple (Malus × domestica cultivar G58) response to infection by the ascomycete fungus Venturia inaequalis causing apple scab disease (Hias et al., 2018). Visual symptoms and PCR quantification of V. inaequalis decreased in the neotetraploid relative to its diploid progenitor. (D) Tetraploid (left) and diploid (right) poker alumroot (Heuchera cylindrica) roots colonized by mycorrhizae fungal arbuscules (the nutritional-exchange structures; Anneberg and Segraves, 2019). Tetraploids show higher colonization rates, as measured by total arbuscules (arrows pointing to arbuscules in each image) than diploids. 200× magnification. Photo credits: (A and B) N. Forrester, (C) N. Hias, and (D) T.J. Anneberg.
Polyploidy may increase quantity or quality aspects of mutualistic associations (Powell and Doyle, 2016; Anneberg and Segraves, 2019; Forrester and Ashman, 2020). For instance, during legume–rhizobium interactions, bacteria fix atmospheric nitrogen into a metabolizable form for the plant in exchange for carbon provided by the plant within root nodules (Figure 2, A and B). Ancient polyploidy is hypothesized to have enhanced this mutualism by increasing the diversity of signaling factors that function in the establishment and maintenance of the interaction in legumes having this symbiosis (Doyle, 2011; Powell and Doyle, 2016, 2017). Changes in root architecture induced by recent polyploidy can also enhance infection rates and increase the total number of nodules produced and number of nitrogen-fixing bacteria hosted (Forrester and Ashman, 2020). Moreover, the “gigas” effect of polyploidy (Stebbins, 1971) can immediately increase nodule size and expand the internal interface for nutrient exchange, as evidenced in synthetic (neo) tetraploid alfalfa subsp. caerulea (Forrester and Ashman, 2020; Figure 2, A and B). Such an effect can increase the efficiency of nitrogen fixation by rhizobial symbionts and thus the amount of nitrogen provided to the plant (Forrester and Ashman, 2020). Lastly, in a test of whether polyploids associate with a wider range of mutualists or gain greater benefits from those with which they do associate, Forrester et al. (2020) exposed diploid and tetraploid alfalfa lineages to a range of bacterial symbionts and found similar niche breadth, but observed that tetraploids attained greater fitness enhancement and maintained this over a larger range of symbionts than diploids.
Root–mycorrhizae associations are widespread and aid plants in obtaining phosphorus, tolerating drought, and provide resistance to root pathogens. Although only recently a focus of study, a positive effect of polyploidy on mycorrhizae colonization has been observed in some cases (Anneberg and Segraves, 2019). In particular, in poker alumroot, tetraploids displayed greater colonization by fungal hyphae and arbuscules, the nutrient exchange structures, than their diploid progenitors (Anneberg and Segraves, 2019; Figure 2D). Although other types of mutualisms (e.g. plant-growth-promoting bacteria in the rhizosphere) are known to assist plant tolerance to stresses such as salt, heavy metals, or pathogens (Rilling et al., 2019) and ploidy-mediated changes in root exudates are predicted to modify these communities (Segraves, 2017), knowledge of how polyploidy affects these processes is lacking. Interestingly, while stress tolerance is often noted as a pathway to invasion for polyploids, potentially accounting for their predominance as invasive species (te Beest et al., 2012; Baduel et al., 2018), another mechanism may be through suppression of the soil microbiome beneficial to natives (Inderjit and Cahill, 2015) via allelopathic root exudates. Whether invasive polyploid plants are especially adept at these remains to be seen. In a 5-year study of diploid and polyploid goldenrod, soil from plots previously inhabited by diploids was significantly different in terms of nutrients, microbes, and soil nematodes relative to soil inhabited by tetraploid or hexaploid plants (Wu et al., 2019), suggesting that polyploidization can affect rhizosphere processes. If these changes in the rhizosphere feed back to benefit the polyploid, they may facilitate polyploid invasion and persistence.
Finally, if the ability of polyploids to tolerate stresses increases through either direct physiological effects or through indirect effects via altered mutualistic interactions or soil biota feedbacks, this increase may change the entire network of species interactions within the community and thus have cascading effects on community structure and resilience (Segraves, 2017). Because communities are the building blocks of ecosystems, and polyploids dominate in some ecoregions more than others (Rice et al., 2019), these changes in stress tolerance may impact nutrient cycling at the level of global biomes. Polyploidy may be selected to enhance mutualisms between animals and other organisms (including those that are not related to abiotic stress). For instance, changes in flower size with polyploidy can lead to more or unique interactions with pollinators (see Roccaforte et al., 2015) that will cascade to affect the structure of the community more broadly. Community-level effects that can be traced to stress response, however, remain another poorly studied area in the broad realm of polyploidy consequences. Nevertheless, the above studies and discussions present intriguing evidence that polyploidy can ameliorate biotic stress, but also illustrate the context-dependency of outcomes of biotic antagonistic and mutualistic interactions.
Stress-induced cell-level polyploidy
Endoreduplication, the result of cell cycle activation without mitosis, leads to tissue-level polyploidy in an otherwise diploid organism (Neiman et al., 2017). Cells that are endopolyploidized often have specific and programmed roles during normal development (Schoenfelder and Fox, 2015; Kolarčik et al., 2020). In plants, well-known examples of specialized endopolyploid cells include fibers and leaf hairs (De Veylder et al., 2011). Endoreduplication has also been proposed to be a plastic response to mitigate the effects of stress (Scholes and Paige, 2015; Bhosale et al., 2018) that may be heritable, and thus a potential target for natural selection. Endoreduplication within plant cells is stimulated by abiotic stresses (e.g. high UV, low temperature, drought, or soil contaminants; Scholes and Paige, 2015) and antagonistic biotic interactions (e.g. disease or herbivory; Smant et al., 2018; Paige, 2019), but also by mutualistic interactions that involve damage to tissues (mycorrhizal fungi and nitrogen-fixing rhizobia, Carotenuto et al., 2019). In animals, endoreduplication is stimulated by damage (e.g. induced wound healing in Drosophila melanogaster epithelium; Losick et al., 2013) and predation (e.g. production of inducible defenses in Daphnia; reviewed in Neiman et al., 2017), but can also be common in secretory or growing tissues (Neiman et al., 2017). Endoreduplication, by increasing cell size, as well as gene copy number and thus gene expression or flux through metabolic pathways, may allow for a rapid response to stress (reviewed in Neiman et al., 2017; Paige, 2019). Notably, in response to various biotic interactions that involve penetration of the plant tissue, endoreduplication results in the upregulation of some host genes that normally regulate cell cycle and cellular hypertrophy, suggesting that the involved effectors are generic reactions to cell stress (Smant et al., 2018). In symbiotic interactions, endoreduplication also functions as a means for differentiation and specialization; for instance, both plant and rhizobial symbionts undergo endoreduplication during nodulation. Indeed, many of the ploidy-related changes in the epigenome in root nodules that house nitrogen-fixing rhizobia are nodule-specific in function (Nagymihaly et al., 2017). Thus, endoreduplication may enhance plant fitness both by mitigating injury and fostering mutualisms, but such plasticity may come with a trade off to whole-organism ploidy (Neiman et al., 2017; Pacey et al., 2020b).
Mechanistically, how can polyploidy mitigate stress?
Physiological and cellular responses to stress
One of the best-known effects of WGD is to increase cell size (Bennett, 1972; Doyle and Coate, 2019). However, the effects of WGD on cell biology are complex and the underlying molecular and physiological mechanisms, as well as the resulting acquired adaptive changes, remain elusive. In some studies, the differences between diploids and polyploids in response to temperature, drought, or salinity stress are attributed to differences in leaf anatomy and/or transpiration rate. For instance, autotetraploid birch (Betula platyphylla) seems better able to maintain water pressure under drought conditions, which is attributed to anatomical differences in leaves. Li et al. observed that polyploids have fewer stomata per unit area, while they also have thicker upper and lower epidermises and more pubescence, which all tend to reduce water loss (Li et al., 1996). Further in birch, Zhang et al. (2016) noticed remarkably different pit-level anatomical traits in polyploids, favoring greater hydraulic safety than their congeneric diploid species and likely contributing to an overabundance of polyploid birch in more stressed habitats. However, the opposite has also been observed. Analysis of leaf anatomy in fireweed (Chamerion angustifolium) revealed morphological adjustments for tolerating water deficiency in diploids in the form of closely packed mesophyll cells and small conduits in the midvein, making diploid C. angustifolium more tolerant to drought than hexaploid plants (Guo et al., 2016).
In Arabidopsis, alterations in cell proliferation and organ size in tetraploids potentiate plant tolerance to salt and drought stresses and decrease transpiration rate, likely through controlling stomatal density and closure, abscisic acid (ABA) signaling, and reactive oxygen species (ROS) homeostasis (del Pozo and Ramirez-Parra, 2014). Indeed, transcriptome deep sequencing (RNA-seq) analysis revealed that the expression of genes involved in redox homeostasis, ABA and stress response was affected (del Pozo and Ramirez-Parra, 2014). In Japanese honeysuckle (Lonicera japonica), water stress decreased net photosynthesis rate, stomatal conductance, and transpiration rate of both diploid and tetraploid cultivars. Water stress also decreased electron transport rate, effective quantum yield of photosystem II, photochemical quenching, and starch content, but increased nonphotochemical quenching and contents for total soluble sugars, proline, and malondialdehyde (Li et al., 2009). However, tetraploid L. japonica showed higher resistance to water stress than diploid species, and physiological responses were less affected in the tetraploid than the diploid. Morphological and anatomical analysis further revealed that the tetraploid had a smaller total plant leaf area, higher leaf mass per unit area, thicker epidermis and palisade tissue, as well as denser pubescence. The suite of specialized structures altered by WGD likely led to greater capacity in dealing with drought stress (Li et al., 2009).
Hexaploid bread wheat (Triticum aestivum) is generally more salt tolerant than its tetraploid progenitor emmer (Triticum turgidum). After allohexaploidization, hexaploid wheat acquired a stronger capacity to retain sodium ions (Na+) within its roots, likely contributing to its higher salt tolerance (Yang et al., 2014a). Similarly, polyploid Arabidopsis accessions are more tolerant to salt stress and better mediate potassium (K+)/Na+ homeostasis under saline conditions than their respective diploid progenitors (Chao et al., 2013). However, the reasons behind this ploidy-determined differential K+/Na+ homeostasis regulation are still largely unknown. Compared to its diploid ancestor, hexaploid morning glory (Ipomoea trifida) efficiently prevents K+ efflux from the meristematic root zone under salt stress through plasma membrane K+ permeable channels, which have low sensitivity to ROS like hydrogen peroxide (H2O2). Moreover, hexaploid I. trifida efficiently excludes Na+ from the elongation and mature root zones under salt stress, because of the high sensitivity of plasma membrane calcium ions (Ca2+) permeable channels to H2O2 (Liu et al., 2019b). However, additional studies are required to unravel the molecular mechanisms underlying this highly cell type-specific ion transport in the roots of hexaploid I. trifida (Liu et al., 2019b).
It remains to be seen to what extent commonalities—with respect to morphological and physiological alterations—exist for different cells and cell types and/or different organisms after polyploidization (Fox et al., 2020). In addition, further studies are necessary to explore the exact molecular and genetic mechanisms shaping condition-dependent morphological and physiological changes in polyploid species.
The genomic response to stress
Long(er) term effects
The reasons for a close relationship between stress and polyploidy have been the subject of considerable speculation, and several hypotheses have been proposed. As stated earlier, most polyploids arise through the formation of unreduced gametes (Ramsey and Schemske, 1998; Mable et al., 2011; Bomblies and Madlung, 2014). However, while increased formation of polyploids in response to stress is one thing, surviving major global change or cataclysmic events is another thing entirely. The K-Pg extinction event likely resulted from a combination of factors, including increased volcanism and in particular the impact of an asteroid near Chicxulub in the Yucatan peninsula (Mexico; Renne et al., 2015). These cataclysmic events ejected dust, sulfate aerosols, and soot particles into the atmosphere, which affected photosynthesis and seed germination during a time of prolonged darkness and dropping temperatures (Vellekoop et al., 2016), resulting in the extinction of up to 70% of all plant species (Nichols and Johnson, 2008). Research suggests that transcription factors with functions in mediating responses to low temperature and shade avoidance, the two main stresses resulting from the K-Pg extinction event, were preferentially retained following WGDs at the K-Pg boundary (Wu et al., 2020). If this finding is correct, the expansion of specific gene families and the resulting dosage effects, or sub- or neofunctionalization of new genes, may have facilitated adaptation to these greatly changed conditions. Biased retention of, in particular, genes with regulatory and developmental functions subsequent to WGDs has been known for many years (Blanc and Wolfe, 2004; Maere et al., 2005) and has been considered by some a mere regulatory or developmental spandrel, a byproduct of the WGD (Freeling, 2009; Birchler and Veitia, 2012). Transcription factor genes, signal transducer genes, developmental genes, and genes encoding proteins that act in multiprotein complexes are retained, not necessarily because of an immediate selective advantage, but mostly because they are important to maintain stoichiometry or gene balance (Freeling, 2009; Birchler and Veitia, 2012; Song et al., 2020). However, given sufficient time, genes can evolve novel functions, facilitating adaptation to the changed environment.
Recent work using an agent-based computational framework built to simulate biological evolution suggested that artificial nonpolyploids perform better than polyploids under stable environments, whereas the opposite holds for unstable or “stressed” environments (Yao et al., 2019, see also Cuypers and Hogeweg, 2014). The evolutionary trajectories of individual genomes in terms of sequence and encoded GRNs were interpreted as follows. Random mutations cause changes in the genome and its associated GRNs. This phenomenon is expected to be amplified in more complex GRNs, such as those arising from WGD, featured by a duplicated number of nodes (i.e. genes or alleles) and an exponential increase in the number of edges (i.e. interactions). Therefore, under stable environments, such random mutations in polyploid genomes will propagate widely throughout the network and will often be maladaptive or detrimental. In contrast, in unstable or stressful environments, these (bigger) changes or disturbances in GRNs may actually provide the substantive changes necessary for survival (Yao et al., 2019; Carretero-Paulet and Van de Peer, 2020).
Duplication of entire genomes and their GRNs immediately creates redundant modules in the genome. Modules are represented by subnetworks within the global GRN that are formed by highly connected genes that are co-expressed and/or co-regulated by the same set of key regulators, while establishing sparser connections with genes from different modules (De Smet and Van de Peer, 2012; Clune et al., 2013; Espinosa-Soto, 2018). There are different ways redundant modules created through WGD may facilitate evolutionary innovation and rapid adaptation to novel environments. First, network redundancy after WGD may help buffer against deleterious mutations, contributing to the genetic or mutational robustness of the organism (Keane et al., 2014). Second, by increasing the absolute dosage of all genes in a pathway (but see Song et al., 2020), WGD can lead to increased fitness under conditions that require elevated fluxes for certain pathways (Van Hoek and Hogeweg, 2009; Bekaert et al., 2011; Hudson et al., 2011; Wu et al., 2020). Third, the probabilities of rewiring biological networks formed by multiple connected genes into specialized metabolic, regulatory, or developmental pathways increase if all involved genes are duplicated together by means of WGD and co-evolve partitioned subfunctions, such as interaction partners or expression domains. This pattern, designated as network subfunctionalization (Blanc and Wolfe, 2004; Conant et al., 2014), has been demonstrated in plants (De Smet et al., 2017). Similarly, modularity increases the probability of rewiring newly duplicated networks into novel biological pathways, a pattern consistent with network neofunctionalization (Sato et al., 2012; Edger et al., 2015; De Smet et al., 2017). The complex and modular structure of duplicated GRNs allow both a wider and faster exploration of phenotypic space and resulting fitness landscapes, ultimately offering increased possibilities for evolutionary innovation and adaptation in a (rapidly) changing environment. Finally, in autopolyploids, each gene is not truly duplicated in a genetic sense, but rather acts as a single locus with up to four alleles segregating within each individual subgenome (tetrasomic inheritance). Although this is different from allopolyploids, the end effects might be very similar. Just as there will be higher gene dosage in autopolyploids because of the additional production of gene products because of the duplication of alleles (Song et al., 2020), there is an increase in the number of edges and nodes, whether between genes or alleles. Of course, one might argue that the effects described above may be different (and perhaps stronger) for allopolyploids, where there are two completely independently evolving loci, and thus less homogenization through recombination, but this needs further study.
Short-term effects
Although some of the processes described above (rewiring of GRNs, sub- or neofunctionalization of genes and networks) take time to exert an effect, genetic variation can also increase immediately upon polyploidization, through the combination of parental genotypes, changes in gene expression, genomic shock, and by epigenetic remodeling (Renny-Byfield and Wendel, 2014; Soltis et al., 2015; Panchy et al., 2016; Van de Peer et al., 2017; Wendel et al., 2018; Qiu et al., 2020). The degree of genomic and transcriptomic changes following WGD is lower in autopolyploids than in allopolyploids, but is highly variable both within and between species in both polyploid types (Wendel, 2015; Spoelhof et al., 2017). Abiotic stress can lead to dramatic changes in the transcriptome of polyploids compared to diploid progenitors (Dong and Adams, 2011; Visger et al., 2019). There is now a vast literature on the influence of genome doubling on gene expression (primarily in allopolyploids, but also in autopolyploids), with most insights garnered via RNA-seq analyses (Chen, 2007; Doyle et al., 2008; Grover et al., 2012; Conant et al., 2014; Yoo et al., 2014; Wendel, 2015; Lian et al., 2020; Nieto Feliner et al., 2020; Qiu et al., 2020). Patterns of gene expression in an allopolyploid can, in part, be the simple addition of its diploid parents. However, many polyploids deviate from the expectations whereby parental gene expression patterns are combined in the allopolyploid. Gene silencing and nonadditive gene up- or down-regulation may occur. A new or young allopolyploid may exhibit novel patterns of gene expression (Grover et al., 2012; Coate et al., 2014; Griffiths et al., 2019), as well as expression level dominance of one parental genome over the other (Yoo et al., 2014; Alger and Edger, 2020). In addition, gene expression differences among duplicate genes present in an allopolyploid are a legacy of expression differences that were already present in the progenitor diploid species (Buggs et al., 2014). In autopolyploid Arabidopsis accessions, a recent study showed that immediate transcriptomic response to genome doubling appears to follow the gene balance hypothesis independently of long-term selective pressures (Song et al., 2020). Overall transcriptome size does not exhibit a simple doubling in response to genome doubling, but instead individual gene dosage responses are highly variable, indicating that expression is not strictly coupled with gene dosage. Nonetheless, putative dosage balance-sensitive gene groups (such as genes encoding multiprotein complexes or genes encoding interacting proteins and metabolic networks) exhibit smaller and more coordinated dosage responses than do putative dosage-insensitive gene groups, suggesting that constraints on dosage balance operate immediately following WGD and that duplicate gene retention patterns are shaped by selection to preserve dosage balance (Song et al., 2020). Also, alternative splicing (AS) has emerged as an important mechanism for generating transcriptomic variation and is very important in stress responses across eukaryote life, including the green lineage (Staiger and Brown, 2013). Significantly, several studies have shown that AS patterns change rapidly following polyploidization and that some or even many of the changes in AS are linked to abiotic stress (Zhang et al., 2010; Zhou et al., 2011; Tack et al., 2014).
Rapid changes in gene expression and epigenetics (see below) following polyploidy may provide the polyploid with an almost immediate selective advantage compared to its diploid progenitors. Indeed, the effects of genome doubling on the phenotype or life history traits can directly affect their chance of survival under stressful conditions. As an example, Godfree et al. determined that tetraploids produced more viable seed and heavier seed than did diploids in kangaroo grass (Themeda triandra), thus increasing fitness and survival under stress (Godfree et al., 2017). More recently, in the same diploid–tetraploid complex, polyploidy was shown to have a direct effect on seed, dormancy, and seedling characteristics (Stevens et al., 2020). Furthermore, tetraploids produced heavier, more viable seed sets that stayed dormant longer under stress, enabling plants to avoid germination under stressful conditions. Furthermore, once they did germinate, tetraploid seedlings were larger and grew faster than did the diploids and were thereby able to outcompete their diploid relatives. Also in T. triandra, it was recently shown that polyploidy appears dominant in hot and dry regions, indicative of ploidy-based adaptive responses to climate (Ahrens et al., 2020). In wheat, higher ploidy levels are associated with higher photosynthetic capacity and stronger morpho–physiological adaptation (Mao et al., 2018). Because polyploid complexes are common in the grasses (Visser and Molofsky, 2015), it would be interesting to know how phenomena such as increased seed viability, seed dormancy, and increased photosynthetic capacity relate to increasing ploidy. Polyploidy has similarly been associated with adaptation to cold stress, facilitating dwarf phenotypes, rapid growth under favorable conditions, and asexual reproduction (te Beest et al., 2012; Freeling, 2017; Syngelaki et al., 2020). It is not difficult to imagine how such traits would provide an advantage under stressful situations, or have increased adaptive potential during or subsequent to cataclysmic events such as those responsible for mass extinction events (Crow and Wagner, 2006).
Stress and epigenetic response
A growing body of evidence points to the importance of epigenetic modifications that accompany WGD. These epigenetic changes can modify homoeologous gene expression patterns, alter networks of gene expression patterns, and may be accompanied by a burst in transposable element (TE) activation. This latter phenomenon has been mainly described for allopolyploids (Parisod et al., 2009; Glombik et al., 2020), although some have reported bursts in TE activation in autopolyploids as well (Bardil et al., 2015; but see Baduel et al., 2019). Ultimately, these changes may facilitate adaptation by new polyploids to their local environment, which includes various stress adaptations (Song and Chen, 2015; Ding and Chen, 2018; Wendel et al., 2018). A study of diploid and autopolyploid alpine buttercup (Ranunculus kuepferi) shows an epigenetic impact of genome doubling on the methylation profile and associated response to cold (Syngelaki et al., 2020). In Arabidopsis hybrids, stress induces the expression of stress–response genes that are normally repressed; the expression of these genes—likely under epigenetic control—allows hybrid plants to combat stress and recover faster than their parents after stress conditions are removed. This role for the timing and operation of stress responses in hybrids is similar to gene expression and growth vigor observed in allopolyploid Arabidopsis (Ni et al., 2009; Miller et al., 2015).
The formation of polyploids significantly increases the complexity of GRNs and transcriptional regulation, which is expected to be reflected in higher-order chromatin structures as well. However, there is little known about three-dimensional (3D) genome structure and its dynamics during polyploidization. By studying 3D genome architectures for both diploid and tetraploid cotton, Wang et al. (2018a) found that by comparing each subgenome in tetraploids with its extant diploid progenitor, genome allopolyploidization contributed to the switching of A/B compartments and the reorganization of topologically associated domains (TADs; Lieberman-Aiden et al., 2009; Lupianez et al., 2015; Dixon et al., 2016). The authors showed that the formation of TAD boundaries during polyploidization preferentially occurred in open chromatin, coinciding with the deposition of active chromatin marks. Furthermore, analysis of inter-subgenomic chromatin interactions revealed the spatial proximity of homoeologous genes, possibly associated with their coordinated expression (Wang et al., 2018a). Although canonical TADs have not been detected in Arabidopsis, hundreds of genomic regions were identified with properties similar to those of TADs (Wang et al., 2015). Allopolyploid Swedish cress (Arabidopsis suecica) showed pronounced expression differences between both parental subgenomes that strongly correlated with chromatin compaction differences between the two homoeologous genomes (Zhu et al., 2017). TAD-like structures have been identified in several additional plant species (Dong et al., 2017), and recent studies have demonstrated the existence of TADs in rice (Dong et al., 2018). Whether analogous compaction differences occur in the homologous genomes of an autopolyploid is as yet unknown (Doyle and Coate, 2019). Likely, there is thus an effect of genome polyploidization on the spatial organization of chromatin, and consequently on gene regulation and gene expression (Jiao et al., 2018; Wang et al., 2018a). To what extent this plays a role in differences between diploids and polyploids, and in their response to stress, needs further work.
What about the proteome?
Although studies of the proteome and the consequences of polyploidy remain in their infancy, it is noteworthy that the proteome does not always mirror the transcriptome (Soltis et al., 2016b). This finding is important in that so much of our current understanding of genetic-based diversity is derived from transcriptome data. In some cases, the polyploid produces qualitative and/or quantitative nonadditive protein patterns compared to the parents of the polyploid, while in others, novel protein production patterns have been observed (Soltis et al., 2016b). Allopolyploids generally combine the protein contributions of their diploid parents, but there is also evidence of one parental genome being dominant. Autopolyploid plants usually are qualitatively identical to their diploid parent, but show quantitative protein differences. Importantly, emerging data show that the proteome and post-translational modifications of proteins play a direct role in the plant immune response (Liu et al., 2019a). For example, Wang et al. (Wang et al., 2013) showed an increased proteomic and physiological response to salt stress in autopolyploid black locust (Robinia pseudoacacia) compared to the diploid progenitor. More studies of the proteome of polyploids and diploid progenitors(s) in response to stress are crucial.
Stress and ecological niche: toward an integration of biotic and abiotic stress response
Given the responses of polyploids to both abiotic and biotic stresses at the cellular and/or organismal levels, it seems likely that responses to stresses might manifest at the landscape scale. Indeed, long standing views on polyploids, especially allopolyploids, suggest that they may have broader ecological amplitude than their diploid progenitors. For example, the higher frequency of polyploids in the Arctic and alpine areas relative to the temperate regions of the Northern Hemisphere was interpreted as evidence for greater stress tolerance, broader ecological niches and possibly even greater colonizing ability in polyploids compared to diploids (e.g. Ehrendorfer, 1980). Although these distributions might also be tied to a higher frequency of formation in habitats in the Arctic/alpine areas following deglaciation (due to increased frequency of unreduced gamete formation in harsh environments and/or novel contact between previously separated diploid species; e.g. Stebbins, 1950, 1984; Novikova et al., 2018; Zozomová-Lihová et al., 2020), latitudinal and elevational gradients in polyploid frequency persist and suggest that polyploids may indeed be better able to cope with harsh environments (Abbott and Brochmann, 2003; Brochmann et al., 2004; Rice et al., 2019). Moreover, ecological divergence from the parental species is generally considered a prerequisite for establishment of a nascent polyploid (Levin, 1975; Fowler and Levin, 1984). Although limited, studies of autopolyploids suggest that the polyploid may escape minority cytotype exclusion through ecological divergence in microhabitat, phenology, pollinators, parasites, or mycorrhizae (Ramsey and Ramsey, 2014; Marchant et al., 2016; Segraves, 2017); even fewer such fine-scale empirical analyses have been reported for allopolyploids.
A polyploid may exhibit ecological niche conservatism with its parental species or it may diverge and exhibit niche expansion, niche contraction, niche intermediacy, or niche novelty (Castro et al., 2020). The application of ecological niche models to the study of polyploids provides a glimpse into the effect of abiotic stresses on both auto- and allopolyploids at the landscape scale. Models for a polyploid and its diploid progenitor(s) generally demonstrate extensive niche overlap between the polyploid and its parent(s), whether an auto- or allopolyploid (Glennon et al., 2012, 2014; Godsoe et al., 2013; Theodoridis et al., 2013; Marchant et al., 2016; Visger et al., 2016; Gaynor et al., 2018), but many polyploids also tend to diverge at least slightly from one or both parents as well. Of course, at present, ecological niche models for plants generally incorporate only abiotic features of climate, elevation, and perhaps soil and land cover, but we can aspire toward incorporating biotic components of the environment as well, from microbiomes to pollinators to herbivores and more. Such models would integrate biotic and abiotic stressors across space and time and thereby permit macroecological and macroevolutionary perspectives on stress response in polyploids. We encourage further study to evaluate more examples of polyploids and their parents to address the relative role of biotic and abiotic stresses in shaping polyploid distributions (Harbert et al., 2014; Rice et al., 2019; Baniaga et al., 2020).
Conclusions
Both individual species, as well as entire ecosystems, can experience stressful conditions; individual plants can be attacked by herbivores or pathogens, while cataclysmic events and/or rapid global change can devastate entire communities. While the effects of all sorts of stress, whether biotic or abiotic, (particularly) on plants are widely documented—see the many excellent reviews on this topic (Erb and Reymond, 2019; Wilkinson et al., 2019; Zhang et al., 2019; Kerchev et al., 2020; Van zelm et al., 2020)—the potential for polyploidy as a stress-tolerance enhancer is much less widely recognized (Fox et al., 2020) and has been based largely on anecdotal evidence (Madlung, 2013; Van de Peer et al., 2017; Doyle and Coate, 2019; Yao et al., 2019; Stevens et al., 2020). Nevertheless, the notion that polyploidy can facilitate response to both abiotic and biotic stresses and that WGD can act as a buffer to mitigate their effects (Van de Peer et al., 2017) is increasingly gaining support. The circumstances and the extent to which polyploidy might confer a selective advantage under stressful conditions, however, as well as the mechanisms underlying the responses to stress-related phenomena, might differ greatly and clearly need further study (Fox et al., 2020).
In a stable environment, gradual changes can successfully explain the divergence and optimization of evolutionary processes. In contrast, when the environment drastically changes, in a short geological time frame, gradual evolution may not keep pace, and existing species may run out of time to adapt and will disappear. Polyploidy might provide a way out of these periods of rapid environmental change and intense stress by enabling swift adaptation to a rapidly changing environment (Crow and Wagner, 2006; Van de Peer et al., 2009; Levin and Soltis, 2018). Gene and genome duplications have been suggested as possible explanations underlying so-called saltational jumps in evolution, consisting of many sudden mutations from one generation to the next (Soltis et al., 2014b, 2016a; Soltis and Soltis, 2014). Indeed, the duplication of genes and particularly the duplication of entire genomes immediately create redundant informational entities or modules in the genome, offering possibilities for a more substantial change and a wider exploration of genotypic and phenotypic space (Conant and Wolfe, 2006; Yao et al., 2019; Carretero-Paulet and Van de Peer, 2020). In a constant environment, organisms are likely to be fairly well-adapted, undergoing only further slow optimization over time; in a changing environment, polyploidy provides a mechanism for more extensive changes. Although such polyploid individuals might initially have difficulty competing with their diploid progenitors, for reasons explained elsewhere (Comai, 2005; te Beest et al., 2012; Van de Peer et al., 2017; Baduel et al., 2018), they could “take their chances” under different conditions or in different contexts. Increased genomic and genetic variation evoked by WGD thus forms a fertile substrate allowing evolution to explore more diverse options, possibly explaining those bigger jumps sometimes observed in evolution. Again, such evolvability (see Payne and Wagner, 2019) might only be successful when the existing environmental conditions have been (seriously) disrupted. Once new environmental conditions have stabilized, the increased evolvability through WGD will become less important in further optimizing the system, and selection on a more complex system might again be constrained. Considering polyploidy in the evolutionary history of organisms might thus indeed be one additional way of explaining large leaps or major transitions that have occurred in the past, as suggested by Susumu Ohno 50 years ago (Ohno, 1970).
Acknowledgements
All authors greatly acknowledge the constructive comments of three anonymous reviewers.
Funding
Y.Vd.P. acknowledges funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant no. 833522). T.L.A. acknowledges funding from the National Science Foundation (grant nos. DEB-1452386 and DEB-1912180).
Conflict of interest statement. None declared.
Contributor Information
Yves Van de Peer, Department of Plant Biotechnology and Bioinformatics, Ghent University, VIB - UGent Center for Plant Systems Biology, B-9052 Ghent, Belgium; Department of Biochemistry, Genetics and Microbiology, University of Pretoria, Pretoria, South Africa; College of Horticulture, Nanjing Agricultural University, Nanjing, China.
Tia-Lynn Ashman, Department of Biological Sciences, University of Pittsburgh, Pittsburgh, Pennsylvania 15260.
Pamela S Soltis, Florida Museum of Natural History, University of Florida, Gainesville, Florida 32611.
Douglas E Soltis, Florida Museum of Natural History, University of Florida, Gainesville, Florida 32611; Department of Biology, University of Florida, Gainesville, Florida 32611.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is Yves Van de Peer (yves.vandepeer@psb.ugent.be).
References
- Abbott RJ, Brochmann C (2003) History and evolution of the arctic flora: in the footsteps of Eric Hulten. Mol Ecol 12: 299–313 [DOI] [PubMed] [Google Scholar]
- Acuna-Rodriguez IS, Newsham KK, Gundel PE, Torres-Diaz C, Molina-Montenegro MA (2020) Functional roles of microbial symbionts in plant cold tolerance. Ecol Lett 23: 1034–1048 [DOI] [PubMed] [Google Scholar]
- Ahrens CW, James EA, Miller AD, Ferguson S, Aitken NC, Jones AW, Lu-Irving P, Borevitz JO, Cantrill DJ, Rymer PD (2020) Spatial, climate, and ploidy factors drive genomic diversity and resilience in the widespread grass Themeda triandra. Mol Ecol 29: 3872–3888 [DOI] [PubMed] [Google Scholar]
- Albuquerque UP, do Nascimentoa ALB, da Silva Chaves L, Soares Feitosa I, de Moura JMB, Gonçalves PHS, da Silva RH, da Silva TC, Soares Ferreira W (2019) A brief introduction to niche construction theory for ecologists and conservationists. Biol Conserv 237: 50–56 [Google Scholar]
- Alger EI, Edger PP (2020) One subgenome to rule them all: underlying mechanisms of subgenome dominance. Curr Opin Plant Biol 54: 108–113 [DOI] [PubMed] [Google Scholar]
- Anneberg TJ, Segraves KA (2019) Intraspecific polyploidy correlates with colonization by arbuscular mycorrhizal fungi in Heuchera cylindrica. Am J Bot 106: 894–900 [DOI] [PubMed] [Google Scholar]
- Arrigo N, Barker MS (2012) Rarely successful polyploids and their legacy in plant genomes. Curr Opin Plant Biol 15: 140–146 [DOI] [PubMed] [Google Scholar]
- Baduel P, Bray S, Vallejo-Marin M, Kolář F, Yant L (2018) The “polyploid hop”: shifting challenges and opportunities over the evolutionary lifespan of genome duplications. Front Ecol Evol 6: 117 [Google Scholar]
- Baduel P, Quadrana L, Hunter B, Bomblies K, Colot V (2019) Relaxed purifying selection in autopolyploids drives transposable element over-accumulation which provides variants for local adaptation. Nat Commun 10: 5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniaga AE, Marx HE, Arrigo N, Barker MS (2020) Polyploid plants have faster rates of multivariate niche differentiation than their diploid relatives. Ecol Lett 23: 68–78 [DOI] [PubMed] [Google Scholar]
- Bardil A, Tayalé A, Parisod C (2015) Evolutionary dynamics of retrotransposons following autopolyploidy in the Buckler Mustard species complex. Plant J 82: 621–631 [DOI] [PubMed] [Google Scholar]
- Barker MS, Arrigo N, Baniaga AE, Li Z, Levin DA (2016) On the relative abundance of autopolyploids and allopolyploids. New Phytol 210: 391–398 [DOI] [PubMed] [Google Scholar]
- Batstone RT, Carscadden KA, Afkhami ME, Frederickson ME (2018) Using niche breadth theory to explain generalization in mutualisms. Ecology 99: 1039–1050 [DOI] [PubMed] [Google Scholar]
- Bekaert M, Edger PP, Pires JC, Conant GC (2011) Two-phase resolution of polyploidy in the Arabidopsis metabolic network gives rise to relative and absolute dosage constraints. Plant Cell 23: 1719–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD (1972) Nuclear DNA content and minimum generation time in herbaceous plants. Proc R Soc Lond B 81: 109–135 [DOI] [PubMed] [Google Scholar]
- Bhosale R, Boudolf V, Cuevas F, Lu R, Eekhout T, Hu Z, Van Isterdael G, Lambert GM, Xu F, Nowack MK, et al. (2018) A spatiotemporal DNA endoploidy map of the arabidopsis root reveals roles for the endocycle in root development and stress adaptation. Plant Cell 30: 2330–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler JA, Veitia RA (2012) Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci U S A 109: 14746–14753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blischak PD, Mabry ME, Conant GC, Pires JC (2018). Integrating networks, phylogenomics, and population genomics for the study of polyploidy. Annu Rev Ecol Evol Syst 49: 253–278 [Google Scholar]
- Bomblies K, Madlung A (2014) Polyploidy in the Arabidopsis genus. Chromosome Res 22: 117–134 [DOI] [PubMed] [Google Scholar]
- Bretagnolle F, Thompson JD (1995) Gametes with the somatic chromosome number: mechanisms of their formation and role in the evolution of autopolyploid plants. New Phytol 129: 1–22 [DOI] [PubMed] [Google Scholar]
- Brochmann C, Brysting AK, Alsos IG, Borgen L, Grundt HH, Scheen AC, Elven R (2004) Polyploidy in arctic plants. Biol J Linn Soc 82: 521–536 [Google Scholar]
- Buggs RA, Wendel JF, Doyle JJ, Soltis DE, Soltis PS, Coate JE (2014) The legacy of diploid progenitors in allopolyploid gene expression patterns. Phil Trans Roy Soc B: Biol Sci 369: 20130354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Xi Z, Amorim AM, Sugumaran M, Rest JS, Liu L, Davis CC (2019) Widespread ancient whole-genome duplications in Malpighiales coincide with Eocene global climatic upheaval. New Phytol 221: 565–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, McKain MR, Harkess A, Nelson MN, Dash S, Deyholos MK, Peng Y, Joyce B, Stewart CN Jr, Rolf M, et al. (2015) Multiple polyploidy events in the early radiation of nodulating and nonnodulating legumes. Mol Biol Evol 32: 193–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carotenuto G, Volpe V, Russo G, Politi M, Sciascia I, de Almeida-Engler J, Genre A (2019) Local endoreduplication as a feature of intracellular fungal accommodation in arbuscular mycorrhizas. New Phytol 223: 430–446 [DOI] [PubMed] [Google Scholar]
- Carretero-Paulet L, Van de Peer Y (2020) The evolutionary conundrum of whole genome duplication. Am J Bot 107: 1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro M, Loureiro J, Figueiredo A, Serrano M, Husband BC, Castro S (2020) Different patterns of ecological divergence between two tetraploids and their diploid counterpart in a parapatric linear coastal distribution polyploid complex. Front Plant Sci 11: 676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao DY, Dilkes B, Luo H, Douglas A, Yakubova E, Lahner B, Salt DE (2013) Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science 341: 658–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ (2007) Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58: 377–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clune J, Mouret JB, Lipson H (2013) The evolutionary origins of modularity. Proc Biol Sci 280: 20122863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coate JE, Bar H, Doyle JJ (2014) Extensive translational regulation of gene expression in an allopolyploid (glycine dolichocarpa). Plant Cell 26: 136–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L (2005) The advantages and disadvantages of being polyploid. Nat Rev Genet 6: 836–846 [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH (2006). Functional partitioning of yeast co-expression networks after genome duplication. PLOS Biol 4: 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9: 938–950 [DOI] [PubMed] [Google Scholar]
- Conant GC, Birchler JA, Pires JC (2014) Dosage, duplication, and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time. Curr Opin Plant Biol 19: 91–98 [DOI] [PubMed] [Google Scholar]
- Crow KD, Wagner GP (2006) What is the role of genome duplication in the evolution of complexity and diversity? Mol Biol Evol 23: 887–892 [DOI] [PubMed] [Google Scholar]
- Cuypers TD, Hogeweg P (2014) A synergism between adaptive effects and evolvability drives whole genome duplication to fixation. PLoS Comput Biol 10: e1003547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet R, Van de Peer Y (2012) Redundancy and rewiring of genetic networks following genome-wide duplication events. Curr Opin Plant Biol 15: 168–176 [DOI] [PubMed] [Google Scholar]
- De Smet R, Sabaghian E, Li Z, Saeys Y, Van de Peer Y (2017) Coordinated functional divergence of genes after genome duplication in Arabidopsis thaliana. Plant Cell 29: 2786–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Storme N, Copenhaver GP, Geelen D (2012) Production of diploid male gametes in Arabidopsis by cold-induced destabilization of postmeiotic radial microtubule arrays. Plant Physiol 160: 1808–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Larkin JC, Schnittger A (2011) Molecular control and function of endoreplication in development and physiology. Trends Plant Sci 16: 624–634 [DOI] [PubMed] [Google Scholar]
- Dehal P, Boore JL (2005) Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol 3: e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Ramirez-Parra E (2014) Deciphering the molecular bases for drought tolerance in Arabidopsis autotetraploids. Plant Cell Environ 37: 2722–2737 [DOI] [PubMed] [Google Scholar]
- Deng B, Du W, Liu C, Sun W, Tian S, Dong H (2012) Antioxidant response to drought, cold and nutrient stress in two ploidy levels of tobacco plants: low resource requirement confers polytolerance in polyploids? Plant Growth Regul 66: 37–47 [Google Scholar]
- Ding M, Chen ZJ (2018) Epigenetic perspectives on the evolution and domestication of polyploid plant and crops. Curr Opin Plant Biol 42: 37–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Gorkin DU, Ren B (2016) Chromatin Domains: The Unit of Chromosome Organization. Mol Cell 62: 668–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P, Tu X, Chu PY, Lu P, Zhu N, Grierson D, Du B, Li P, Zhong S (2017) 3D chromatin architecture of large plant genomes determined by local A/B compartments. Mol Plant 10: 1497–1509 [DOI] [PubMed] [Google Scholar]
- Dong Q, Li N, Li X, Yuan Z, Xie D, Wang X, Li J, Yu Y, Wang J, Ding B, et al. (2018) Genome‐wide Hi‐C analysis reveals extensive hierarchical chromatin interactions in rice. Plant J 94: 1141–1156 [DOI] [PubMed] [Google Scholar]
- Dong S, Adams KL (2011) Differential contributions to the transcriptome of duplicated genes in response to abiotic stresses in natural and synthetic polyploids. New Phytol 190: 1045–1057 [DOI] [PubMed] [Google Scholar]
- Doyle JJ (2011) Phylogenetic perspectives on the origins of nodulation. Mol Plant-Microbe Interact 24: 1289–1295 [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Coate JE (2019) Polyploidy, the nucleotype, and novelty: the impact of genome doubling on the biology of the cell. Int J Plant Sci 180: 1–52 [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF (2008) Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet 42: 443–461 [DOI] [PubMed] [Google Scholar]
- Edger PP, Heidel-Fischer HM, Bekaert M, Rota J, Glockner G, Platts AE, Heckel DG, Der JP, Wafula EK, Tang M, et al. (2015) The butterfly plant arms-race escalated by gene and genome duplications. Proc Natl Acad Sci U S A 112: 8362–8366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrendorfer F (1980) Polyploidy and distribution. Plenum Press, New York, USA. [Google Scholar]
- Erb M, Reymond P (2019) Molecular interactions between plants and insect herbivores. Annu Rev Plant Biol 70: 527–557 [DOI] [PubMed] [Google Scholar]
- Espinosa-Soto C (2018) On the role of sparseness in the evolution of modularity in gene regulatory networks. PLoS Comput Biol 14: e1006172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estep MC, McKain MR, Vela Diaz D, Zhong J, Hodge JG, Hodkinson TR, Layton DJ, Malcomber ST, Pasquet R, Kellogg EA (2014) Allopolyploidy, diversification, and the Miocene grassland expansion. Proc Natl Acad Sci U S A 111: 15149–15154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett JA, Maere S, Van de Peer Y (2009) Plants with double genomes might have had a better chance to survive the Cretaceous-Tertiary extinction event. Proc Natl Acad Sci U S A 106: 5737–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folk R, Soltis DE (2020) Angiosperms at the edge: extremity and diversity. Plant Cell Environ 43: 2871–93 [DOI] [PubMed] [Google Scholar]
- Forrester NJ, Ashman TL (2020) Synthetic autotetraploids show that polyploidy alters the mutualism interface of legume-rhizobia interactions in Medicago sativa subsp. caerulea. Am J Bot 107: 179–18331721161 [Google Scholar]
- Forrester NJ, Rebolleda‐Gómez M, Sachs JL, Ashman TL (2020) Polyploid plants obtain greater fitness benefits from a nutrient acquisition mutualism. New Phytol 227: 944–954 [DOI] [PubMed] [Google Scholar]
- Foster CB, Afonin SA (2005) Abnormal pollen grains: an outcome of deteriorating atmospheric conditions around the Permian - Triassic boundary. J Geol Soc London 162: 653–659 [Google Scholar]
- Fowler NL, Levin DA (1984) Ecological constraints on the establishment of a novel polyploid in competition with its diploid progenitor. Am Nat 124: 703–711 [Google Scholar]
- Fox D, Soltis DE, Soltis PS, Ashman T-L, Van de Peer Y (2020) Polyploidy: a biological force from cells to ecosystems. Trends Cell Biol 30: 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M (2009) Bias in plant gene content following different sorts of duplication: tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol 60: 433–453 [DOI] [PubMed] [Google Scholar]
- Freeling M (2017) The distribution of ancient polyploidies in the plant phylogenetic tree is a spandrel of occasional sex. Plant Cell 29: 202–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor ML, Marchant DB, Soltis DE, Soltis PS (2018) Absence of niche divergence among ploidal levels in the classic autopolyploid system, Galax urceolata (Diapensiaceae). Am J Bot 105: 1631–1642 [DOI] [PubMed] [Google Scholar]
- Glennon KL, Rissler LJ, Church SA (2012) Ecogeographic isolation: a reproductive barrier between species and between cytotypes in Houstonia (Rubiaceae). Evol Ecol 26: 909–926 [Google Scholar]
- Glennon KL, Ritchie ME, Segraves KA (2014) Evidence for shared broad-scale climatic niches of diploid and polyploid plants. Ecol Lett 17: 574–582 [DOI] [PubMed] [Google Scholar]
- Glombik M, Bačovský V, Hobza R, Kopecký D (2020) Competition of parental genomes in plant hybrids. Front Plant Sci 11: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godden GT, Kinser TJ, Soltis PS, Soltis DE (2019) Phylotranscriptomic analyses reveal asymmetrical gene duplication dynamics and signatures of ancient polyploidy in mints. Genome Biol Evol 11: 3393–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfree RC, Marshall DJ, Young AG, Miller CH, Mathews S (2017) Empirical evidence of fixed and homeostatic patterns of polyploid advantage in a keystone grass exposed to drought and heat stress. R Soc Open Sci 4: 170934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godsoe W, Larson MA, Glennon KL, Segraves KA (2013) Polyploidization in Heuchera cylindrica (Saxifragaceae) did not result in a shift in climatic requirements. Am J Bot 100: 496–508 [DOI] [PubMed] [Google Scholar]
- Griffiths AG, Moraga R, Tausen M, Gupta V, Bilton TP, Campbell MA, Ashby R, Nagy I, Khan A, Larking A, et al. (2019) Breaking free: the genomics of allopolyploidy-facilitated niche expansion in white clover. Plant Cell 31: 1466–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover CE, Gallagher JP, Szadkowski EP, Yoo MJ, Flagel LE, Wendel JF (2012) Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol 196: 966–971 [DOI] [PubMed] [Google Scholar]
- Gunn BF, Murphy DJ, Walsh NG, Conran JG, Pires JC, Macfarlane TD, Birch JL (2020) Evolution of Lomandroideae: multiple origins of polyploidy and biome occupancy in Australia. Mol Phyl Evol 149: 106836. [DOI] [PubMed] [Google Scholar]
- Guo W, Yang J, Sun XD, Chen GJ, Yang YP, Duan YW (2016) Divergence in eco-physiological responses to drought mirrors the distinct distribution of Chamerion angustifolium cytotypes in the Himalaya-Hengduan mountains region. Front Plant Sci 7: 1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannweg K, Steyn W, Bertling I (2016) In vitro-induced tetraploids of Plectranthus esculentus are nematode-tolerant and have enhanced nutritional value. Euphytica 207: 343–351 [Google Scholar]
- Harbert RS, Brown AHD, Doyle JJ (2014) Climate niche modeling in the perennial Glycine (Leguminosae) allopolyploid complex. Am J Bot 110: 710–721 [DOI] [PubMed] [Google Scholar]
- Harms N, Shearer J, Cronin JT, Gaskin JF (2020) Geographic and genetic variation in susceptibility of Butomus umbellatus to foliar fungal pathogens. Biol Invasions 22: 535–548 [Google Scholar]
- Hias N, Svara A, Keulemans JH (2018) Effect of polyploidisation on the response of apple (Malus × domestica Borkh.) to Venturia inaequalis infection. Eur J Plant Pathol 151: 515–526 [Google Scholar]
- Huang CH, Zhang C, Liu M, Hu Y, Gao T, Qi J, Ma H (2016) Multiple polyploidization events across Asteraceae with two nested events in the early history revealed by nuclear phylogenomics. Mol Biol Evol 33: 2820–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson CM, Puckett EE, Bekaert M, Pires JC, Conant GC (2011) Selection for higher gene copy number after different types of plant gene duplications. Genome Biol Evol 3: 1369–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inderjit, Cahill JL (2015) Linkages of plant–soil feedbacks and underlying invasion mechanisms. AoB Plants 7: plv022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao W, Yuan J, Jiang S, Liu Y, Wang L, Liu M, Zheng D, Ye W, Wang X, Chen ZJ (2018) Asymmetrical changes of gene expression, small RNAs and chromatin in two resynthesized wheat allotetraploids. Plant J 93: 828–842 [DOI] [PubMed] [Google Scholar]
- Kagale S, Robinson SJ, Nixon J, Xiao R, Huebert T, Condie J, Kessler D, Clarke WE, Edger PP, Links MG, et al. (2014) Polyploid evolution of the Brassicaceae during the Cenozoic era. Plant Cell 26: 2777–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keane OM, Toft C, Carretero-Paulet L, Jones GW, Fares MA (2014) Preservation of genetic and regulatory robustness in ancient gene duplicates of Saccharomyces cerevisiae. Genome Res 24: 1830–1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchev P, van der Meer T, Sujeeth N, Verlee A, Stevens CV, Van Breusegem F, Gechev T (2020) Molecular priming as an approach to induce tolerance against abiotic and oxidative stresses in crop plants. Biotechnol Adv 40: 107503. [DOI] [PubMed] [Google Scholar]
- Koenen EJM, Ojeda DI, Bakker FT, Wieringa JJ, Kidner C, Hardy OJ, Pennington RT, Herendeen PS, Bruneau A, Hughes CE (2020) The origin of the legumes is a complex paleopolyploid phylogenomic tangle closely associated with the cretaceous–paleogene (K–Pg) mass extinction event. Syst Biol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolarčik V, Fráková V, Kocová V, Koprivý L, Mártonfi P (2020) Endopolyploidy pattern in Corydalis early spring geophytes. Flora 270: 151651 [Google Scholar]
- Kurschner WM, Batenburg SJ, Mander L (2013) Aberrant Classopollis pollen reveals evidence for unreduced (2n) pollen in the conifer family Cheirolepidiaceae during the Triassic-Jurassic transition. Proc Biol Sci 280: 20131708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis JB, Soltis DE, Li Z, Marx HE, Barker MS, Tank DC, Soltis PS (2018) Impact of whole- genome duplication events on diversification rates in angiosperms. Am J Bot 105: 348–363 [DOI] [PubMed] [Google Scholar]
- Laport RG, Ng J (2017) Out of one, many: the biodiversity considerations of polyploidy. Am J Bot 104: 1119–1121 [DOI] [PubMed] [Google Scholar]
- Le Comber SC, Smith CP (2004) Polyploidy in fishes: patterns and processes. Eur J Linnean Soc 82: 431–442 [Google Scholar]
- Leebens-Mack JH, Barker MS, Carpenter EJ, Deyholos MK, Gitzendanner MA, Graham SW, Grosse I, Li Z, Melkonian M, Mirarab S, et al. (2019) One thousand plant transcriptomes and the phylogenomics of green plants. Nature 574: 679–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin DA (1975) Minority cytotype exclusion in local plant populations. Taxon 24: 35–43 [Google Scholar]
- Levin DA (1983) Polyploidy and novelty in flowering plants. Am Nat 122: 1–25 [Google Scholar]
- Levin DA, Soltis DE (2018) Factors promoting polyploid persistence and diversification and limiting diploid speciation during the K–Pg interlude. Curr Opin Plant Biol 42: 1–7 [DOI] [PubMed] [Google Scholar]
- Li W-D, Biswas DK, Xu H, Xu C-Q, Wang X-Z, Liu J-K, Jiang G-M (2009) Photosynthetic responses to chromosome doubling in relation to leaf anatomy in Lonicera japonica subjected to water stress. Funct Plant Biol 36: 783–792 [DOI] [PubMed] [Google Scholar]
- Li W-L, Berlyn GP, Ashton PMS (1996) Polyploids and their structural and physiological characteristics relative to water deficit in Betula papyrifera (Betulaceae). Am J Bot 83: 15–20 [Google Scholar]
- Li X, Guo B (2020) Substantially adaptive potential in polyploid cyprinid fishes: evidence from biogeographic, phylogenetic and genomic studies. Proc Biol Sci 287: 20193008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Shen H, Zhou Q, Qian K, van der Lee T, Huang S (2017) Changing ploidy as a strategy: the Irish potato famine pathogen shifts ploidy in relation to its sexuality. Mol Plant Microbe Interact 30: 45–52 [DOI] [PubMed] [Google Scholar]
- Lian S, Zhou Y, Liu Z, Gong A, Cheng L (2020) The differential expression patterns of paralogs in response to stresses indicate expression and sequence divergences. BMC Plant Biol 20: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Ren Y, Li Z, Hu Q, Yin L, Qiao X, Zhang Y, Xing L, Xi Y, Jiang F, et al. (2020) Giant African snail genomes provide insights into molluscan whole-genome duplication and aquatic-terrestrial transition. Mol Ecol Resour (in press) [DOI] [PubMed] [Google Scholar]
- Liu Y, Lu S, Liu K, Wang S, Huang L, Guo L (2019a) Proteomics: a powerful tool to study plant responses to biotic stress. Plant Methods 15: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yu Y, Sun J, Cao Q, Tang Z, Liu M, Xu T, Ma D, Li Z, Sun J (2019b) Root-zone-specific sensitivity of K+-and Ca2+-permeable channels to H2O2 determines ion homeostasis in salinized diploid and hexaploid Ipomoea trifida. J Exp Bot 70: 1389–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick VP, Fox DT, Spradling AC (2013) Polyploidization and cell fusion contribute to wound healing in the adult Drosophila epithelium. Curr Biol 23: 2224–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourkisti R, Froelicher Y, Herbette S, Morillon R, Tomi F, Gibernau M, Giannettini J, Berti L, Santini J (2020) Triploid citrus genotypes have a better tolerance to natural chilling conditions of photosynthetic capacities and specific leaf volatile organic compounds. Front Plant Sci 11: 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. (2015) Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mable BK, Alexandrou MA, Taylor MI (2011) Genome duplication in amphibians and fish: an extended synthesis. J Zool 284: 151–182 [Google Scholar]
- Madlung A (2013) Polyploidy and its effect on evolutionary success: old questions revisited with new tools. Heredity 110: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A, Tyagi AP, Watson B, Jiang HM, Kagochi T, Doerge RW, Martienssen R, Comai L (2005) Genomic changes in synthetic Arabidopsis polyploids. Plant J 41: 221–230 [DOI] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y (2005) Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci U S A 102: 5454–5459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali H, Walden AE, Husband BC (2009) Genome duplication and the evolution of physiological responses to water stress. New Phytol 184: 721–731 [DOI] [PubMed] [Google Scholar]
- Mandakova T, Lysak MA (2018) Post-polyploid diploidization and diversification through dysploid changes. Curr Opin Plant Biol 42: 55–65 [DOI] [PubMed] [Google Scholar]
- Mao H, Chen M, Su Y, Wu N, Yuan M, Yuan S, Brestic M, Zivcak M, Zhang H, Chen Y (2018) Comparison on photosynthesis and antioxidant defense systems in wheat with different ploidy levels and octoploid triticale. Int J Mol Sci 19: 3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchant BD, Soltis DE, Soltis PS (2016) Patterns of abiotic niche shifts in allopolyploids relative to their progenitors. New Phytol 212: 708–718 [DOI] [PubMed] [Google Scholar]
- Mason AS, Nelson MN, Yan G, Cowling WA (2011) Production of viable male unreduced gametes in Brassica interspecific hybrids is genotype specific and stimulated by cold temperatures. BMC Plant Biol 11: 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M, Song Q, Shi X, Juenger TE, Chen ZJ (2015) Natural variation in timing of stress-responsive gene expression predicts heterosis in intraspecific hybrids of Arabidopsis. Nat Comm 6: 7453. [DOI] [PubMed] [Google Scholar]
- Moore LJ, Nilsen TO, Jarungsriapisit J, Fjelldal PG, Stefansson SO, Taranger GL, Patel S (2017) Triploid atlantic salmon (Salmo salar L.) post-smolts accumulate prevalence more slowly than diploid salmon following bath challenge with salmonid alphavirus subtype 3. PLoS One 12: e0175468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan C, Zhang H, Henry CE, Franklin FCH, Bomblies K (2020) Derived alleles of two axis proteins affect meiotic traits in autotetraploid Arabidopsis arenosa. Proc Natl Acad Sci U S A 17: 8980–8988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagymihaly M, Veluchamy A, Gyorgypal Z, Ariel F, Jegu T, Benhamed M, Szucs A, Kereszt A, Mergaert P, Kondorosi E (2017) Ploidy-dependent changes in the epigenome of symbiotic cells correlate with specific patterns of gene expression. Proc Natl Acad Sci U S A 114: 4543–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman M, Beaton MJ, Hessen DO, Jeyasingh PD, Weider LJ (2017) Endopolyploidy as a potential driver of animal ecology and evolution. Biol Rev 92: 234–247 [DOI] [PubMed] [Google Scholar]
- Ni DA, Sozzani R, Blanchet S, Domenichini S, Reuzeau C, Cella R, Bergounioux C, Raynaud C (2009) The Arabidopsis MCM2 gene is essential to embryo development and its over-expression alters root meristem function. New Phytol 184: 311–322 [DOI] [PubMed] [Google Scholar]
- Nichols DJ, Johnson KR (2008) Plants and the K-T Boundary. Cambridge University Press, Cambridge, UK. [Google Scholar]
- Nieto Feliner G, Casacuberta J, Wendel JF (2020) Genomics of evolutionary novelty in hybrids and polyploids. Front Genet 11: 792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikova PY, Hohmann N, Van de Peer Y (2018) Polyploid Arabidopsis species originated around recent glaciation maxima. Curr Opin Plant Biol 42: 8–15 [DOI] [PubMed] [Google Scholar]
- Novikova PY, Brennan IG, Booker W, Mahony M, Doughty P, Lemmon AR, Lemmon EM, Roberts JD, Yant L, Van de Peer Y, et al. (2020) Polyploidy breaks speciation barriers in Australian burrowing frogs Neobatrachus. PLoS Genet (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlander KC, Dreyer LL, Goldblatt P, Suda J, Linder HP (2016) Species-rich and polyploid-poor: Insights into the evolutionary role of whole-genome duplication from the Cape flora biodiversity hotspot. Am J Bot 103: 1336–1347 [DOI] [PubMed] [Google Scholar]
- Ohno S (1970) Evolution by Gene Duplication. Springer Verlag, New York [Google Scholar]
- Oswald BP, Nuismer SL (2007) Neopolyploidy and pathogen resistance. P Roy Soc B Biol Sci 274: 2393–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacey EK, Maherali H, Husband BC (2020a) Endopolyploidy is associated with leaf functional traits and climate variation in Arabidopsis thaliana. Am J Bot 107: 993–1003 [DOI] [PubMed] [Google Scholar]
- Pacey EK, Maherali H, Husband BC (2020b) The influence of experimentally induced polyploidy on the relationships between endopolyploidy and plant function in Arabidopsis thaliana. Ecol Evol 10: 198–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige KN (2019) Overcompensation, environmental stress, and the role of endoreduplication. Am J Bot 105: 1–4 [DOI] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu S-H (2016) Evolution of gene duplication in plants. Plant Physiol 171: 2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisod C, Salmon A, Zerjal T, Tenaillon M, Grandbastien M-A, Ainouche M (2009) Rapid structural and epigenetic reorganization near transposable elements in hybrid and allopolyploid genomes in Spartina. New Phytol 184: 1003–1015 [DOI] [PubMed] [Google Scholar]
- Payne JL, Wagner A (2019) The causes of evolvability and their evolution. Nat Rev Genet 20: 24–38 [DOI] [PubMed] [Google Scholar]
- Pecrix Y, Rallo G, Folzer H, Cigna M, Gudin S, Le Bris M (2011) Polyploidization mechanisms: temperature environment can induce diploid gamete formation in Rosa sp. J Exp Bot 62: 3587–3597 [DOI] [PubMed] [Google Scholar]
- Peters MJ, Suwannapong G, Pelin A, Corradi N (2019) Genetic and genome analyses reveal genetically distinct populations of the bee pathogen Nosema ceranae from Thailand. Microb Ecol 77: 877–889 [DOI] [PubMed] [Google Scholar]
- Powell AF, Doyle JJ (2016) Enhanced rhizobial symbiotic capacity in an allopolyploid species of Glycine (Leguminosae). Am J Bot 103: 1771–1782 [DOI] [PubMed] [Google Scholar]
- Powell AF, Doyle JJ (2017) Non-additive transcriptomic responses to inoculation with rhizobia in a young allopolyploid compared with its diploid progenitors. Genes 8: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu T, Liu Z, Liu B (2020) The effects of hybridization and genome doubling in plant evolution via allopolyploidy. Mol Biol Rep 47: 5549–5558 [DOI] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst 29: 467–501 [Google Scholar]
- Ramsey J, Ramsey TS (2014) Ecological studies of polyploidy in the 100 years following its discovery. Phil Trans R Soc B: Biol Sci 369: 20130352–20130352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne PR, Sprain CJ, Richards MA, Self S, Vanderkluysen L, Pande K (2015) State shift in Deccan volcanism at the Cretaceous-Paleogene boundary, possibly induced by impact. Science 350: 76–78 [DOI] [PubMed] [Google Scholar]
- Renny‐Byfield S, Wendel JF (2014) Doubling down on genomes: polyploidy and crop plants. Am J Bot 101: 1711–1725 [DOI] [PubMed] [Google Scholar]
- Rice A, Šmarda P, Novosolov M, Drori M, Glick L, Sabath N, Meiri S, Belmaker J, Mayrose I (2019) The global biogeography of polyploid plants. Nature Ecol Evol 3: 265–273 [DOI] [PubMed] [Google Scholar]
- Rilling JI, Acuña JJ, Nannipieri P, Cassan F, Maruyama F, Jorquera MA (2019) Current opinion and perspectives on the methods for tracking and monitoring plant growth‒promoting bacteria Soil Biol Biochem 130: 205–219 [Google Scholar]
- Robertson FM, Gundappa MK, Grammes F, Hvidsten TR, Redmond AK, Lien S, Martin SAM, Holland PWH, Sandve SR, Macqueen DJ (2017) Lineage-specific rediploidization is a mechanism to explain time-lags between genome duplication and evolutionary diversification. Genome Biol 18: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roccaforte K, Russo SE, Pilson D (2015) Hybridization and reproductive isolation between diploid Erythronium mesochoreum and its tetraploid congener E. albidum (Liliaceae). Evolution 69: 1375–1389 [DOI] [PubMed] [Google Scholar]
- Ruiz M, Quinones A, Martinez-Cuenca MR, Aleza P, Morillon R, Navarro L, Primo-Millo E, Martinez-Alcantara B (2016) Tetraploidy enhances the ability to exclude chloride from leaves in carrizo citrange seedlings. J Plant Physiol 205: 1–10 [DOI] [PubMed] [Google Scholar]
- Sato S, Tabata S, Hirakawa H, Asamizu E, Shirasawa K, Isobe S, Kaneko T, Nakamura Y, Shibata D, Aoki K, et al. (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder KP, Fox DT (2015) The expanding implications of polyploidy. J Cell Biol 209: 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes DR, Paige KN (2015) Plasticity in ploidy: a generalized stress response. Trends Plant Science 20: 165–175 [DOI] [PubMed] [Google Scholar]
- Schranz ME, Mohammadin S, Edger PP (2012) Ancient whole genome duplications, novelty and diversification: the WGD Radiation Lag-Time Model. Curr Opin Plant Biol 15: 147–153 [DOI] [PubMed] [Google Scholar]
- Segraves KA (2017) The effects of genome duplications in a community context. New Phytol 215: 57–69 [DOI] [PubMed] [Google Scholar]
- Selmecki AM, Maruvka YE, Richmond PA, Guillet M, Shoresh N, Sorenson AL, De S, Kishony R, Michor F, Dowell R, et al. (2015) Polyploidy can drive rapid adaptation in yeast. Nature 519: 349–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa EB (2019) Polyploidy as a mechanism for surviving global change. New Phytol 221: 5–6 [DOI] [PubMed] [Google Scholar]
- Smant G, Helder J, Goverse A (2018) Parallel adaptations and common host cell responses enabling feeding of obligate and facultative plant parasitic nematodes. Plant J 93: 686–702 [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS (1999) Polyploidy: recurrent formation and genome evolution. Trends Ecol Evol 14: 348–352 [DOI] [PubMed] [Google Scholar]
- Soltis DE, Misra BB, Shan S, Chen S, Soltis PS (2016b) Polyploidy and the proteome. Biochim Biophys Acta 1864: 896–907 [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Schemske D, Hancock J, Thompson J, Husband B, Judd WS (2007) Autopolyploidy and sympatric speciation in angiosperms: have we grossly underestimated the number of species? Taxon 56: 13–30 [Google Scholar]
- Soltis DE, Visger CJ, Marchant DB, Soltis PS (2016a) Polyploidy: Pitfalls and paths to a paradigm. Am J Bot 103: 1146–1166 [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE (2009) The role of hybridization in plant speciation. Annu Rev Plant Biol 60: 561–588 [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE (2014) Flower diversity and angiosperm diversification. Methods Mol Biol 1110: 85–102 [DOI] [PubMed] [Google Scholar]
- Soltis PS, Marchant DB, Van de Peer Y, Soltis DE (2015) Polyploidy and genome evolution in plants. Curr Opin Genet Dev 35: 119–125 [DOI] [PubMed] [Google Scholar]
- Soltis DE, Visger CJ, Soltis PS (2014a) The polyploidy revolution then … and now: Stebbins revisited. Am J Bot 101: 1057–1078 [DOI] [PubMed] [Google Scholar]
- Soltis PS, Liu X, Marchant DB, Visger CJ, Soltis DE (2014b) Polyploidy and novelty: Gottlieb's legacy. Philos Trans R Soc Lond B Biol Sci 369: 20130351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MJ, Potter BI, Doyle JJ, Coate JE (2020) Gene balance predicts transcriptional responses immediately following ploidy change in Arabidopsis thaliana. Plant Cell 32: 1434–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Chen ZJ (2015) Epigenetic and developmental regulation in plant polyploids. Curr Opin Plant Biol 24: 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoelhof JP, Soltis PS, Soltis DE (2017) Pure polyploidy: closing the gaps in autopolyploid research: pure polyploidy. J Syt Evol 55: 340–352 [Google Scholar]
- Staiger D, Brown JW (2013) Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25: 3640–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL (1971) Chromosomal Evolution in Higher Plants. Edward Arnold LTD, London, pp. 87–89 [Google Scholar]
- Stebbins GL (1984) Polyploidy and the distribution of the Arctic-Alpine Flora - new evidence and a new approach. Bot Helv 94: 1–13 [Google Scholar]
- Stebbins GL Jr (1950) Variation and Evolution in Plants. Columbia University Press, New York. [Google Scholar]
- Stebbins GL Jr (1949) The evolutionary significance of natural and artificial polyploids in the family Gramineae. Hereditas 35: 461–485 [Google Scholar]
- Stevens AV, Nicotra AB, Godfree RC, Guja LK (2020) Polyploidy affects the seed, dormancy and seedling characteristics of a perennial grass, conferring an advantage in stressful climates. Plant Biol 22: 500–513 [DOI] [PubMed] [Google Scholar]
- Syngelaki E, Schinkel CCF, Klatt S, Hörandl E (2020) Effects of temperature treatments on cytosine-methylation profiles of diploid and autotetraploid plants of the Alpine species Ranunculus kuepferi (Ranunculaceae). Front Plant Sci 11: 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tack DC, Pitchers WR, Adams KL (2014) Transcriptome analysis indicates considerable divergence in alternative splicing between duplicated genes in Arabidopsis thaliana. Genetics 198: 1473–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Raes J (2004) Duplication and divergence: the evolution of new genes and old ideas. Annu Rev Genet 38: 615–643 [DOI] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y (2003) Genome duplication, a trait shared by 22000 species of ray-finned fish. Genome Res 13: 382–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, Kubesova M, Pysek P (2012) The more the better? The role of polyploidy in facilitating plant invasions. Ann Bot 109: 19–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoridis S, Randin C, Broennimann O, Patsiou T, Conti E (2013) Divergent and narrower climatic niches characterize polyploid species of European primroses in Primula sect. Aleuritia. J Biogeogr 40: 1278–1289 [Google Scholar]
- Thompson JD, Lumaret R (1992) The evolutionary dynamics of polyploid plants: origins, establishment and persistence. Trends Ecol Evol 7: 302–307 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y (2004) Computational approaches to unveiling ancient genome duplications. Nat Rev Genet 5: 752–763 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A (2009) The evolutionary significance of ancient genome duplications. Nat Rev Genet 10: 725–732 [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Mizrachi E, Marchal K (2017) The evolutionary significance of polyploidy. Nat Rev Genet 18: 411–424 [DOI] [PubMed] [Google Scholar]
- Van Hoek MJA, Hogeweg P (2009) Metabolic adaptation after whole genome duplication. Mol Biol Evol 26: 2441–2453 [DOI] [PubMed] [Google Scholar]
- Van zelm E, Zhang Y, Testerink C (2020) Salt tolerance mechanisms of plants. Annu Rev Plant Biol 71: 403–433 [DOI] [PubMed] [Google Scholar]
- Vanneste K, Baele G, Maere S, Van de Peer Y (2014) Analysis of 41 plant genomes supports a wave of successful genome duplications in association with the Cretaceous-Paleogene boundary. Genome Res 24: 1334–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellekoop J, Esmeray-Senlet S, Miller KG, Browning JV, Sluijs A, van de Schootbrugge B, Sinninghe Damsté JS, Brinkhuis H (2016) Evidence for Cretaceous-Paleogene boundary bolide “impact winter” conditions from New Jersey, USA. Geology 44: 619–622 [Google Scholar]
- Visger CJ, Wong GK, Zhang Y, Soltis PS, Soltis DE (2019) Divergent gene expression levels between diploid and autotetraploid Tolmiea relative to the total transcriptome, the cell, and biomass. Am J Bot 106: 280–291 [DOI] [PubMed] [Google Scholar]
- Visger CJ, Germain-Aubrey CC, Patel M, Sessa EB, Soltis PS, Soltis DE (2016) Niche divergence between diploid and autotetraploid Tolmiea. Am J Bot 103: 1396–1406 [DOI] [PubMed] [Google Scholar]
- Visscher H, Looy CV, Collinson ME, Brinkhuis H, van Konijnenburg-Van Cittert JHA, Kurschner WM, Sephton MA (2004) Environmental mutagenesis during the end-Permian ecological crisis. Proc Natl Acad Sci USA 101: 12952–12956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser V, Molofsky J (2015) Ecological niche differentiation of polyploidization is not supported by environmental differences among species in a cosmopolitan grass genus. Am J Bot 102: 36–49 [DOI] [PubMed] [Google Scholar]
- Wang C, Liu C, Roqueiro D, Grimm D, Schwab R, Becker C, Lanz C, Weigel D (2015) Genome-wide analysis of local chromatin packing in Arabidopsis thaliana. Genome Res 25: 246–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang P, Lin M, Ye Z, Li G, Tu L, Shen C, Li J, Yang Q, Zhang X (2018a) Evolutionary dynamics of 3D genome architecture following polyploidization in cotton. Nat Plants 4: 90–97 [DOI] [PubMed] [Google Scholar]
- Wang W, He Y, Cao Z, Deng Z (2018b) Induction of tetraploids in Impatiens (Impatiens walleriana) and characterization of their changes in morphology and resistance to downy mildew. HortScience 53: 925–931 [Google Scholar]
- Wang Z, Wang M, Liu L, Meng F (2013) Physiological and proteomic responses of diploid and tetraploid black locust (Robinia pseudoacacia L.) subjected to salt stress. Int J Mol Sci 2013 14: 20299–20325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF (2015) The wondrous cycles of polyploidy in plants. Am J Bot 102: 1753–1756 [DOI] [PubMed] [Google Scholar]
- Wendel JF, Lisch D, Hu G, Mason AS (2018) The long and short of doubling down: polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr Opin Genet Devel 49: 1–7 [DOI] [PubMed] [Google Scholar]
- Wilkinson SW, Magerøy MH, López Sánchez A, Smith LM, Furci L, Cotton TEA, Krokene P, Ton J (2019) Surviving in a hostile world: plant strategies to resist pests and diseases. Annu Rev Plant Biol 57: 505–529 [DOI] [PubMed] [Google Scholar]
- Wolfe KH (2001) Yesterday's polyploids and the mystery of diploidization. Nature Rev Genet 2: 333–341 [DOI] [PubMed] [Google Scholar]
- Wong GK, Soltis DE, Leebens-Mack J, Wickett NJ, Barker MS, Van de Peer Y, Graham SW, Melkonian M (2020) Sequencing and analyzing the transcriptomes of a thousand species across the tree of life for green plants. Annu Rev Plant Biol 71: 1.1–1.25 [DOI] [PubMed] [Google Scholar]
- Wu S, Han B, Jiao Y (2020) Genetic contribution of paleopolyploidy to adaptive evolution in angiosperms. Mol Plant 13: 59–71 [DOI] [PubMed] [Google Scholar]
- Wu S, Cheng J, Xu X, Zhang Y, Zhao Y, Li H, Qiang S (2019) Polyploidy in invasive Solidago canadensis increased plant nitrogen uptake, and abundance and activity of microbes and nematodes in soil. Soil Biol Biochem 138: 10794 [Google Scholar]
- Yang C, Zhao L, Zhang H, Yang Z, Wang H, Wen S, Zhang C, Rustgi S, von Wettstein D, Liu B (2014a) Evolution of physiological responses to salt stress in hexaploid wheat. Proc Natl Acad Sci U S A 111: 11882–11887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PM, Huang QC, Qin GY, Zhao SP, Zhou JG (2014b) Different drought-stress responses in photosynthesis and reactive oxygen metabolism between autotetraploid and diploid rice. Photosynthetica 52: 193–202 [Google Scholar]
- Yao Y, Carretero-Paulet L, Van de Peer Y (2019) Using digital organisms to study the evolutionary consequences of whole genome duplication and polyploidy. PLoS One 14: e0220257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo MJ, Liu X, Pires JC, Soltis PS, Soltis DE (2014) Nonadditive gene expression in polyploids. Annu Rev Genet 48: 485–517 [DOI] [PubMed] [Google Scholar]
- Zedek F, Plackova K, Vesely P, Smerda J, Smarda P, Horova L, Bures P (2020) Endopolyploidy is a common response to UV-B stress in natural plant populations, but its magnitude may be affected by chromosome type. Ann Bot 126: 883–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li X-M, Lin HX, Chong K (2019) Crop improvement through temperature resilience. Annu Rev Plant Biol 70: 753–780 [DOI] [PubMed] [Google Scholar]
- Zhang PG, Huang SZ, Pin AL, Adams KL (2010) Extensive divergence in alternative splicing patterns after gene and genome duplication during the evolutionary history of Arabidopsis. Mol Biol Evol 27: 1686–1697 [DOI] [PubMed] [Google Scholar]
- Zhang W-W, Song J, Wang M, Liu Y-Y, Li N, Zhang Y-J, Michele Holbrook N, Hao G-Y (2016) Divergences in hydraulic architecture form an important basis for niche differentiation between diploid and polyploid Betula species in NE China. Tree Physiol 37: 604–616 [DOI] [PubMed] [Google Scholar]
- Zhou R, Moshgabadi N, Adams KL (2011) Extensive changes to alternative splicing patterns following allopolyploidy in natural and resynthesized polyploids. Proc Natl Acad Sci U S A 108: 16122–16127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Zhao S, Lu X, He N, Gao L, Dou J, Bie Z, Liu W (2018) Genome duplication improves the resistance of watermelon root to salt stress. Plant Physiol Biochem 133: 11–21 [DOI] [PubMed] [Google Scholar]
- Zhu W, Hu B, Becker C, Doğan ES, Berendzen KW, Weigel D, Liu C (2017) Altered chromatin compaction and histone methylation drive non-additive gene expression in an interspecific Arabidopsis hybrid. Genome Biol 18: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zozomová-Lihová J, Melichárková A, Svitok M, Španiel S (2020) Pleistocene range disruption and postglacial expansion with secondary contacts explain the genetic and cytotype structure in the western Balkan endemic Alyssum austrodalmaticum (Brassicaceae). Plant Syst Evol 306: 47 [Google Scholar]