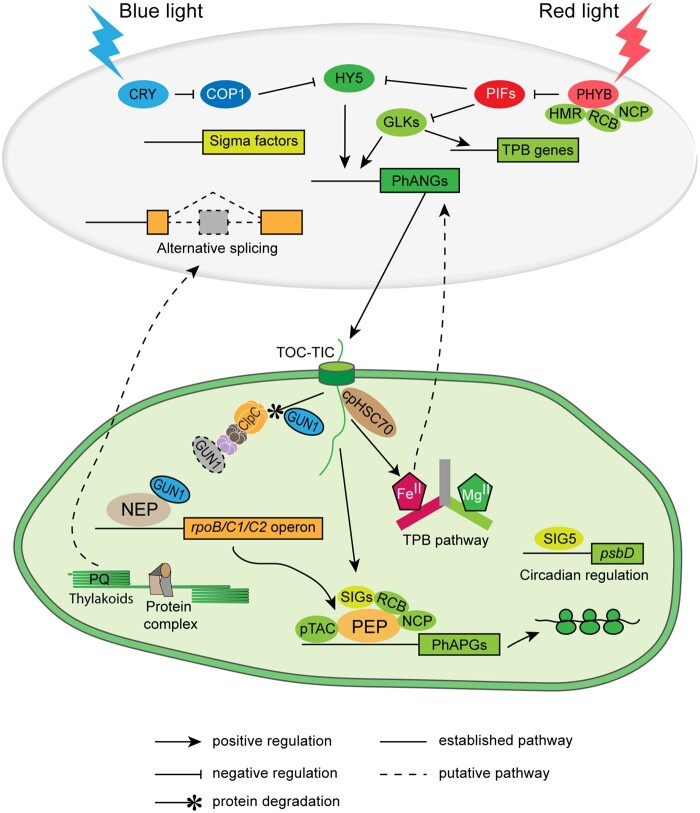
Figure 3.
Interaction of light and plastid signals during chloroplast biogenesis. In the dark, active plastid-encoded genes are mainly transcribed by NEP. GUN1 was reported to directly bind NEP and promote the expression of NEP-dependent genes upon inhibited plastid translation (Tadini et al., 2020). Upon illumination, the activated form of blue-light and red-light photoreceptors (CRY and phy) degrade negative regulators (COP1 and PIFs, respectively) of photomorphogenesis, thus activating the expression of positive transcriptional regulators (e.g. HY5 and GLKs), thereby promoting the expression of PhANGs and TPB genes. The PhANG-encoded proteins and TPB enzymes are imported into plastids (proplastids, etioplasts, and etiochloroplasts) through the TOC-TIC apparatus. GUN1 facilitates protein import during chloroplast biogenesis, and is degraded by the Clp protease when chloroplast biogenesis is complete. PEP is the major RNA polymerase in chloroplasts, and its activity requires the plastid-encoded core components RpoA, RpoB, RpoC1, and RpoC2, and nucleus-encoded accessory factors, including bacterial-type Sigma factors (SIGs) for promoter recognition. Other nucleus-encoded factors support complex assembly (e.g. pTACs [PAP8, HMR], REGULATOR OF CHLOROPLAST BIOGENESIS [RCB], and NUCLEAR CONTROL OF PEP ACTIVITY [NCP]) and regulate PEP activity, for example, PLASTID REDOX INSENSITIVE2 (PRIN2) that is involved in redox regulation of PEP activity (Díaz et al., 2018). The nucleus-encoded Sigma factor SIG5 communicates circadian information from the nucleus to the chloroplast and controls expression of at least 12% of all chloroplast-encoded genes (Noordally et al., 2013). Chloroplast-derived signals promote PhANG expression through positive signaling molecules such as heme, regulate alternative splicing of transcripts from nuclear genes (through the redox status of the plastoquinone pool, PQ; Petrillo et al., 2014), and facilitate nuclear body-localization of the red-light photoreceptor phyB and the degradation of negative regulators (PIFs; Chen et al., 2010; Galvao et al., 2012; Yang et al., 2019; Yoo et al., 2019).