Figure 9.
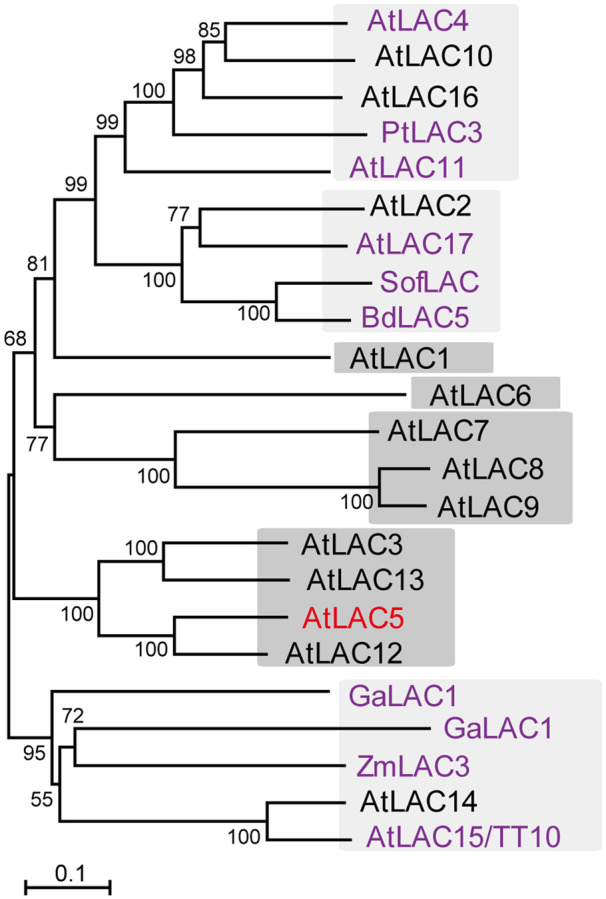
Nonrooted molecular phylogenetic tree of Arabidopsis LACs and functionally identified LACs. Functionally identified LACs are shown in purple. The clusters with unknown LACs only are shown with a dark grey background. The tree was constructed as described in Materials and methods section. Bar = 0.1 amino acid substitutions per site. The GenBank accession numbers for the sequences are shown in parentheses: AtLAC1 (At1g18140, NP_173252); AtLAC2 (At2g29130, NP_180477); AtLAC3 (At2g30210, NP_180580); AtLAC4 (At2g38380, NP_565881); AtLAC5 (At2g40370, NP_181568); AtLAC6 (At46570, NP_182180); AtLAC7 (At3g09220, NP_187533); AtLAC8 (At5g01040, NP_195724); AtLAC9 (At5g01050, NP_195725); AtLAC10 (At5g01190, NP_195739); AtLAC11 (At5g03260, NP_195946); AtLAC12 (At5g05390, NP_196158); AtLAC13 (At5g07130, NP_196330); AtLAC14 (At5g09360, NP_196498); AtLAC15 (At5g48100, NP_199621); AtLAC16 (At5g58910, NP_200699); AtLAC17 (At5g60020, NP_200810); BdLAC5 (Bradi1g66720, XP_003558240); BnTT10 (NP_001302959); GaLAC1 (NP_001316972); PtLAC3 (CAC14719); SofLAC (SCVPRZ3027A08.g and SCUTST3084C11.g); ZmLAC3 (CAJ30499). At, Arabidopsis thaliana; Bd, Brachypodium distachyon; Bn, Brassica napus; Ga, Gossypium arboreum; Pt, Populus trichocarpa; Sof, Saccharum officinarum; Zm, Zea mays. The alignment used for this analysis is shown in Supplemental Figure 3.
