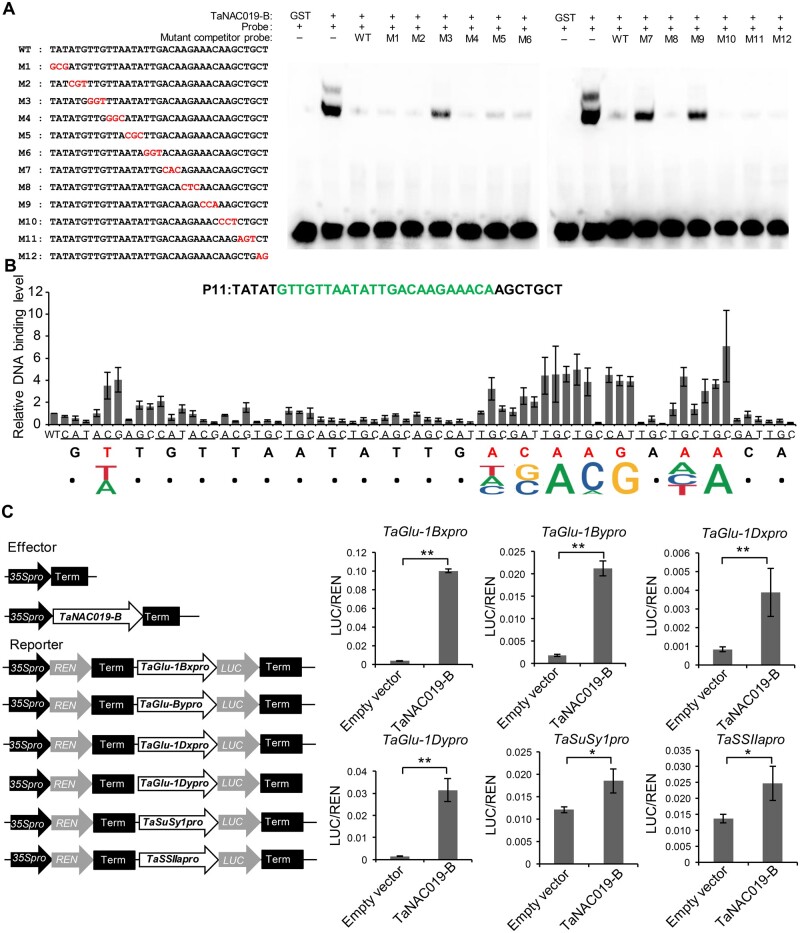
Figure 4.
TaNAC019 binds to a specific motif in the TaGlu promoters. A, A three-nucleotide mutation series spanning the P11 oligonucleotide sequence was generated (highlighted in red) and used as competitors of the P11 probe in EMSAs with TaNAC019-B-GST recombinant protein. “+” and “−” indicate the presence and absence, respectively, of the corresponding probes and proteins. B, Mononucleotide mutations were introduced into the P11 core region (highlighted in green) and used as competitor probes in EMSAs. Each nucleotide of the core region was converted into the three other nucleotides. The average values of TaNAC019-B-binding levels from three independent replicates are shown on the y-axis. Nucleotides required for TaNAC019-B binding are indicated in red and the deduced binding motif is shown along the x-axis. C, Dual luciferase transcriptional activity assay to assess the ability of TaNAC019-B to transactivate target gene expression. Schematic diagrams of the effector and reporter plasmids. On the y-axis, LUC/REN indicates the ratio of the signal detected for firefly luciferase (LUC) versus Renilla reniformis luciferase (REN) activity. Error bars represent standard deviation for three replicates. The single and double asterisks indicate statistically significant differences estimated by Student’s t test at P < 0.05 and P < 0.01, respectively.