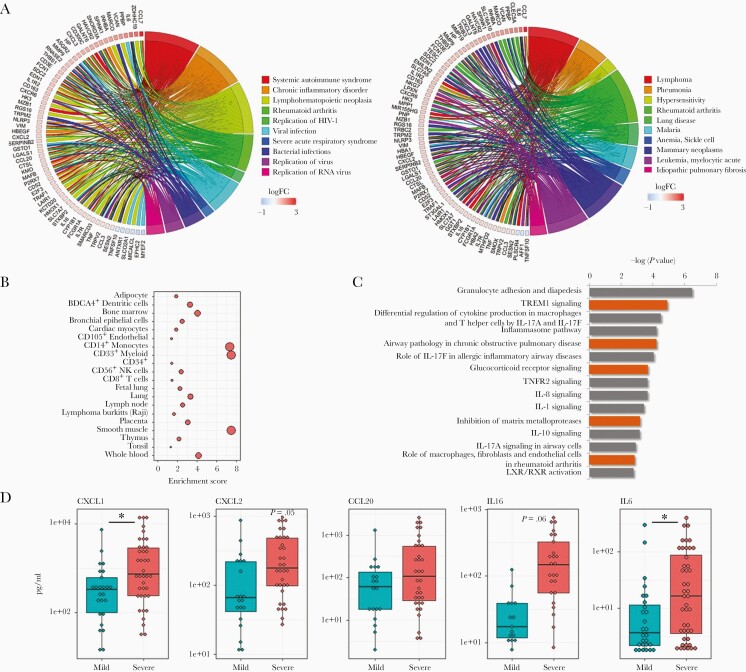
Figure 1.
Airway gene expression patterns associated with clinical severity in RSV-infected infants. Gene expression from nasal/airway samples, collected from 106 infants infected with RSV, was assessed by high-throughput RNA sequencing. A, Circos plots displaying biological function (left) and disease (right) ontologies associated with severity. Shown are a subset of genes with expression patterns significantly associated (FDR < .05, fold change > 1.5) with clinical severity, following adjustment for other variables in a multivariate model, and key ontologies significantly associated with severity (n = 629 genes, FDR < .05). Biological functions are enriched in viral infection-related ontologies, and disease ontologies include pneumonia, a lower airway pathology. B, Cell type enrichment plots indicate severity genes are associated with multiple hematopoietic lineages, predominantly CD14+ monocytes, CD33+ myeloid cells, and smooth muscle cells. C, Canonical pathway analysis of severity-associated genes suggests alterations in immune signaling in infants with severe clinical symptoms, including IL-17. D, Multiplex ELISA analysis of nasal washes from RSV-infected infants confirms that the production of select immune-related proteins are increased in infants with severe clinical symptoms. Asterisk, P value < .05; Boxes, upper and lower quartiles of data; Whiskers, the highest and lowest data point. Abbreviations: ELISA, enzyme-linked immunosorbent assay; FDR, false discovery rate; IL-17, interleukin-17; RSV, respiratory syncytial virus.