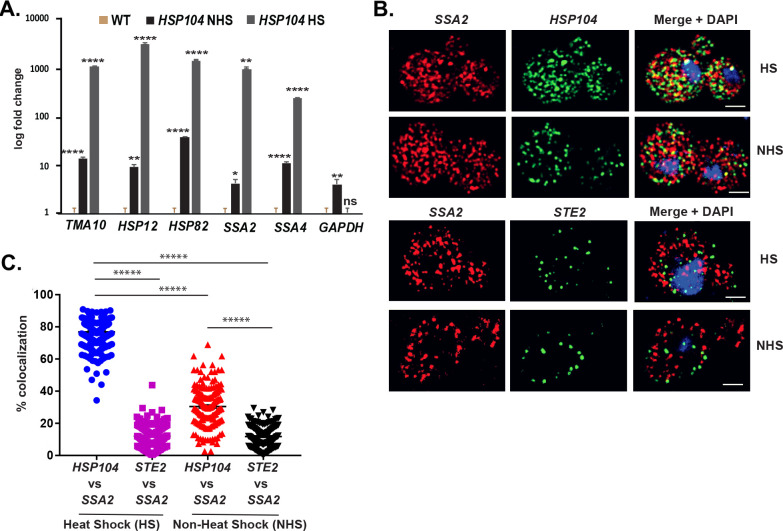
Figure 8. mRNAs encoding heat shock proteins undergo enhanced multiplexing upon heat shock.
MATa yeast strains (BY4741) expressing MS2 aptamer-tagged HSP104, UGO1, or STE5 from their genomic loci or untagged control (WT) cells were grown to mid-log phase (O.D.600 = 0.5) and subjected to RaPID followed by qRT-PCR (RaPID-qRTPCR; see 'Materials and methods'). RNA derived from the total cell extracts or biotin-eluated fractions was analyzed by qRT-PCR using primer pairs corresponding to mRNAs expected to multiplex (see listed genes) or not (e.g., GAPDH mRNA). (A) mRNAs encoding heat shock proteins (HSPs) multiplex upon heat shock. Cells expressing MS2 aptamer-tagged HSP104 were either exposed to heat shock (10 min; 40°C; HS) or maintained at 30°C (NHS) prior to fixation and RaPID-qRT-PCR. Three biological replicates were performed and an unpaired t-test was used to compare each under non-heat and heat shock conditions with WT; ****p<0.0001; **p<0.001; *p<0.01. (B) mRNAs of HSPs co-localize upon heat shock. Representative single-molecule fluorescence in situ hybridization (smFISH) images of BY4741 WT cells that underwent heat shock (HS) or did not (NHS). BY4741 were processed for smFISH labeling using designated FISH probes complementary to the HSP104 and SSA2 and HSP104 and STE2 mRNA pairs prior to labeling with DAPI. SSA2 – Cy3 labeling; HSP104/STE2 – Alexa488 labeling; merge – merged Cy3 and Alexa-488 windows with DAPI staining. Size bar = 2 μm. The representative image shown is from a single focal plane. (C) Scatter plot of data from (B) showing the proportion of co-localized smFISH foci. Black lines represent average ± SEM distribution. Each data point represents a single cell. *****p-value<0.00001.