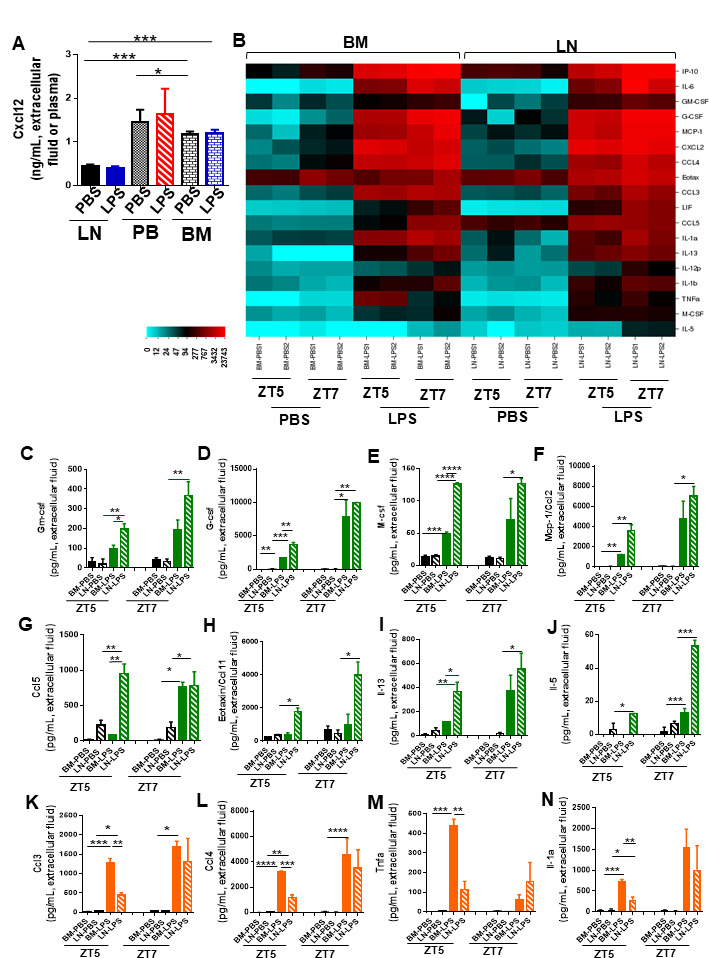
Figure 4. Traf6 is a key regulator for migration of bone marrow (BM)-derived myeloid progenitors to lymph nodes (LNs) in a non-cell-autonomous manner.
(A) Schema of full chimeric mice made by non-competitive transplantation of CD45.2+ Mx1Cre;WT and Mx1Cre;Traf6flox/flox BM cells into lethally irradiated CD45.1+ B6.SJLPtprca Pep3b/BoyJ. Six weeks later Traf6 gene were deleted by intraperitoneal injection of poly(I:C). 1 week later we performed PBS/lipopolysaccharide (LPS) injection early in the rest phase (zeitgeber time [ZT]4 [10 am, time of LPS administration]) and LN-contained myeloid progenitors at ZT7 (1 pm, 3 hr after LPS administration) was scored by colony-forming unit (CFU) assay. (B) Absolute number of CFU-GM present in LN from wild-type (WT) (solid bars) and Traf6∆/∆ (orange bars) full chimeric mice (n = 6–7 mice per group) after PBS (black and orange solid bars) or LPS (blue and mosaic bars) administration. (C–H) In vitro transwell migration assay for BM-derived LK cells. (C) Experimental design for migration of WT or Traf6∆/∆ low-density (LD) BM cells (CD45.2+) toward a WT microenvironment generated by BM (CD45.1+) in the presence of LPS for 4 hr. (D) Graph represents the percentage migrated LK from WT (blue solid bar) or Traf6∆/∆ (orange mosaic bar) low-density BM (LDBM) cells to the bottom as depicted in (C). (E) Experimental design for migration of WT LDBM cells (CD45.1+) toward a gradient generated by WT or Traf6∆/∆ (CD45.2+) BM or LN cells in the presence of LPS for 4 hr. (F) Graph represents the percentage of LDBM LK migrated to the BM bottom (solid bars) or LN bottom (mosaic bars) as schemed in (E). (G) Experimental design for WT LDBM cells (CD45.1+) migration toward gradient generated by WT or Traf6∆/∆ LN-derived T-cells (CD45.2+/CD3e+/CD11b-/B220-) or B-cells (CD45.2+/CD3e-CD11b-/B220+) or myeloid cells (CD45.2+/CD3e-/CD11b+/B220-) in the presence of LPS for 4 hr. (H) Graph represents the percentage of migrated LK LDBM to the WT LN bottom (blue mosaic bars) or Traf6∆/∆ LN bottom (orange mosaic bars) as schemed in (G). In all cases, LK cell migration was determined by CD45 allotype analysis using flow cytometry in triplicate. (I) Absolute number of CFU-GM present in LN from Lyz2Cre;WT (solid bars) and Lyz2Cre;Traf6flox/flox (mosaic bars) full chimeric mice after PBS/LPS administration at ZT7 (3 hr). (J) Graph represents cumulative survival of Lyz2Cre;WT (blue line) and Lyz2Cre;Traf6flox/flox (orange line) after 10 mg/kg of LPS. (J) Survival curve after 30 mg/kg of b.w. injection in Lyz2Cre;WT (blue line) or Lyz2Cre;Traf6flox/flox (orange line). Values are shown as mean ± SD of two independent experiments with a minimum of three mice or replicates per group and experiment. *p<0.05, **p<0.01.
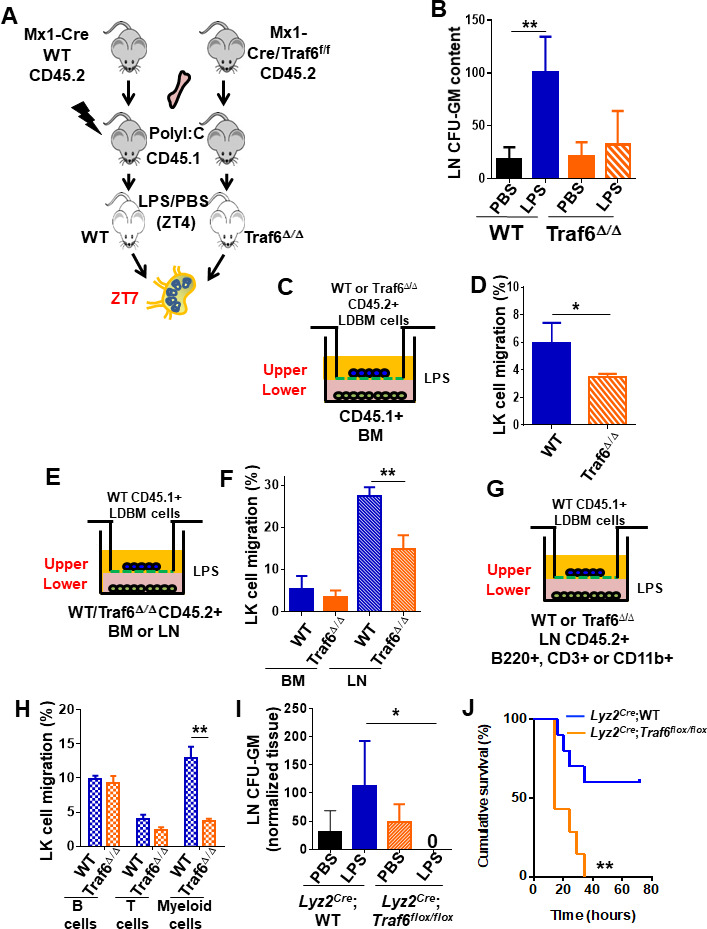
Figure 4—figure supplement 1. Myeloid progenitor migration to lymph node (LN) in response to lipopolysaccharide (LPS) is independent of NF-κB activation.
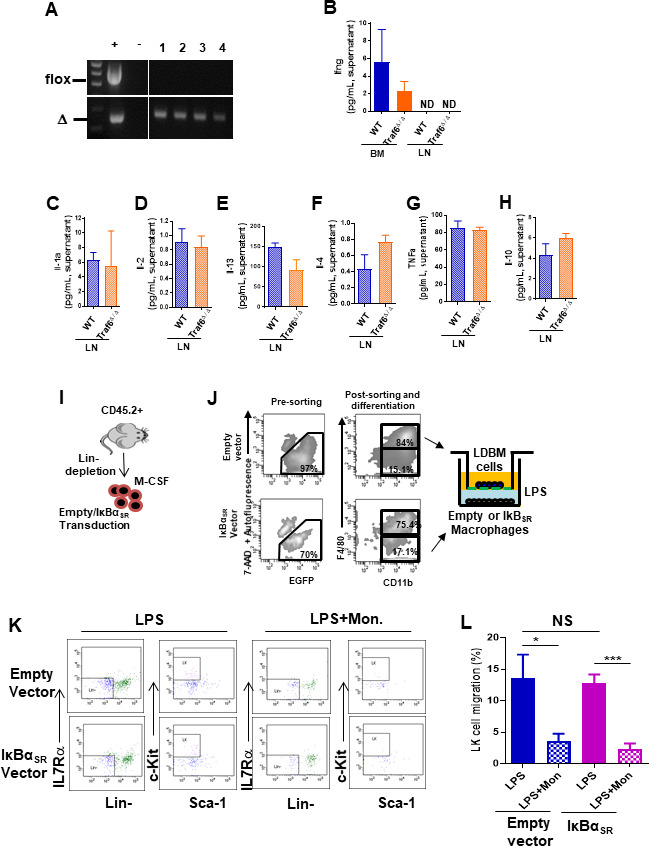
Figure 4—figure supplement 2. Inflammation induces temporal changes in chemokine and cytokine signatures in bone marrow (BM) and lymph node (LN).