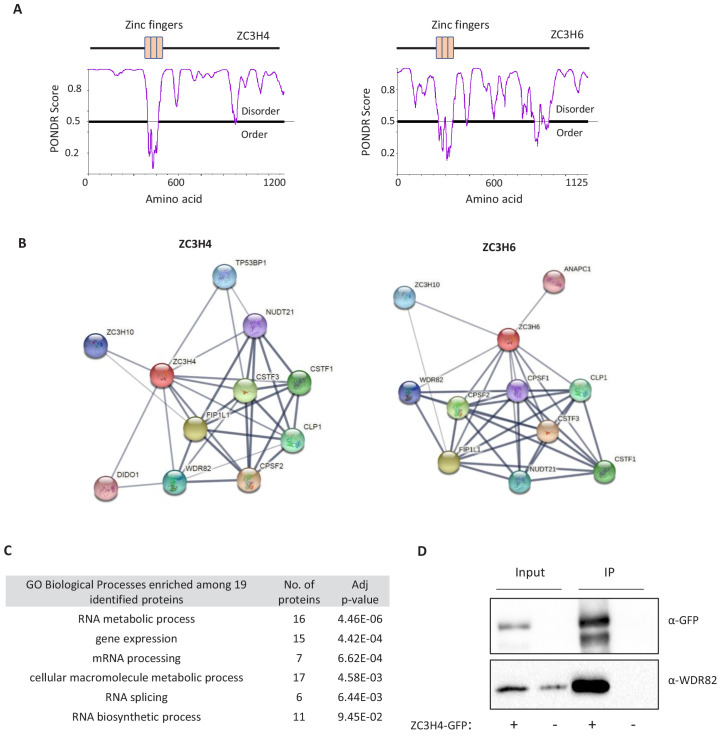
Figure 1. Proximity labelling of CPSF30-sensitive Pol II interactions by mTurbo.
(a) Schematic of the strategy used to tag CPSF30 with the mini auxin-inducible degron (mAID). Guide RNA-expressing Cas9 plasmid and homology-directed repair (HDR) plasmids are shown and the resulting modification to CPSF30 is represented with each inserted element labelled. (b) Western blot demonstrating CPSF30 depletion. Parental HCT116-TIR1, or CPSF30-mAID cells, were treated ±auxin for 3 hr, then blotted. CPSF30 protein is indicated together with a non-specific product, marked by an asterisk, used as a proxy for protein loading. (c) Metagene analysis of 1795 protein-coding genes demonstrating increased downstream transcription, derived from sequencing nuclear RNA, following auxin treatment (3 hr) of CPSF30-mAID cells. TSS = transcription start site, TES = transcription end site (PAS), read-through signal is normalised against gene body. RPKM is reads per kilobase of transcript, per million mapped reads. Positive and negative signals represent sense and antisense reads, respectively. (d) Schematic of our strategy to identify new factors involved in transcription termination. CPSF30-mAID cells were edited to express Rpb1-mTurbo (blue circle on Pol II). The addition of biotin induces mTurbo-mediated biotinylation (orange haze) of factors proximal to Pol II. CPSF complex is shown as an example of what might be captured by this experiment. (e) Western blot showing streptavidin horseradish peroxidase (HRP) probing of extracts from CPSF30-mAID: RPB1-mTurbo cells. Prior treatment with auxin (3 hr)/biotin (10 min) is indicated. The high molecular weight species in the +biotin samples corresponds in size to Rpb1-mTurbo (*). (f) Heat map detailing proteins with the largest decrease in Pol II interaction. Data underpinning heat map are from mass spectrometry analysis of streptavidin sequestered peptides (±CPSF30) performed in triplicate. Labelling was for 10 min.