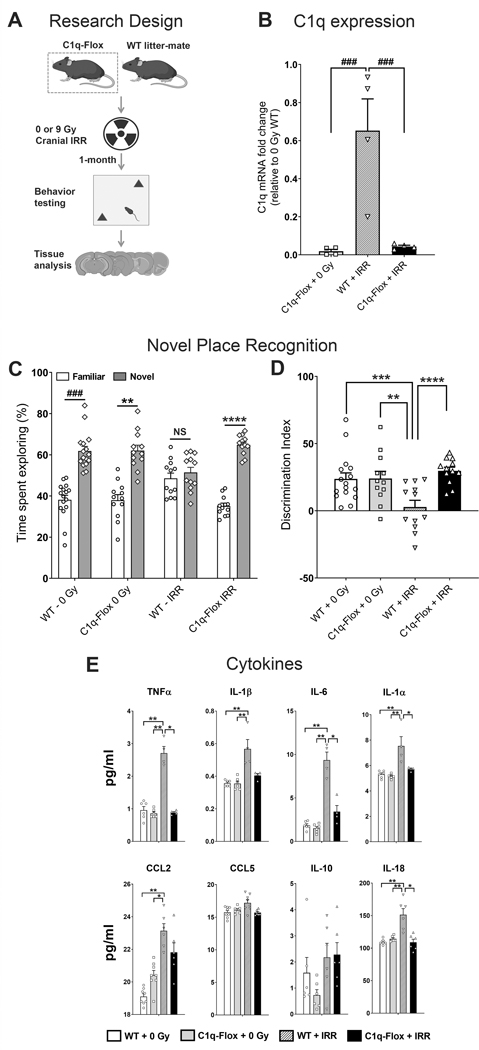
Figure 5. Microglia-selective deletion of complement C1q protect against radiation-induced cognitive impairments and elevated pro-inflammatory cytokines.
(A) Four month old C1q-Flox (C1qaFL/FL:Cx3cr1CreERT2) and littermate control (WT) mice were exposed to 0 or 9 Gy cranial irradiation and one month post-irradiation mice were administered hippocampal-dependent novel place recognition task. (B) Quantification of C1q expression (qPCR) in vivo showed a significant reductions (96–98%, P’s = 0.0001) in the C1q-Flox 0 and 9 Gy irradiated brains. (C) Irradiated (IRR) WT mice spent significantly less time exploring novel spatial location. Time spent exploring the novel placements of objects during the test phase of the novel place recognition task show that 0 Gy WT, 0 Gy C1q-Flox and irradiated C1q-Flox mice spent significantly more time exploring novel versus familiar location whereas irradiated WT mice spent comparable time exploring both locations. (D) The tendency to explore novel spatial location was derived from the Discrimination Index, calculated as ([Novel location exploration time/Total exploration time] – [Familiar location exploration time/Total exploration time]) × 100. Cranial irradiation significantly impaired cognitive function in the WT mice as shown by the reduced preference towards the novel place compared to the 0 Gy WT, 0 Gy C1q-Flox and irradiated C1q-Flox mice (P = 0.01, P= 0.02 and P = 0.001 respectively). Importantly, irradiated C1q-Flox mice did not show a decline on the spatial exploration. (E) Multiplex quantification of cytokines show radiation-induced elevations in the pro-inflammatory factors TNFα, IL-1β, IL-6, IL-1α, CCL2 and IL-18 in the WT-IRR brains whereas irradiated C1q-Flox mice did not show increases in the cytokine levels. Data are presented as mean ± SEM (N = 4 observations per group, B; N = 16 observations per group, C-D; N = 3 to 5 observations per group, E). P values were derived from Kruskal-Wallis test (B and E), Wilcoxon matched-pairs signed rank test (C) and, two-way ANOVA and Bonferroni’s post hoc test (D). NS, not significant; *, P = 0.03; **, P=0.02, ***, P =0.01; ****, P = 0.001; ### P = 0.0001.