Figure 3. Amino acid substitutions in the FWRs of the scFv 763.74(A) destabilize the scFv.
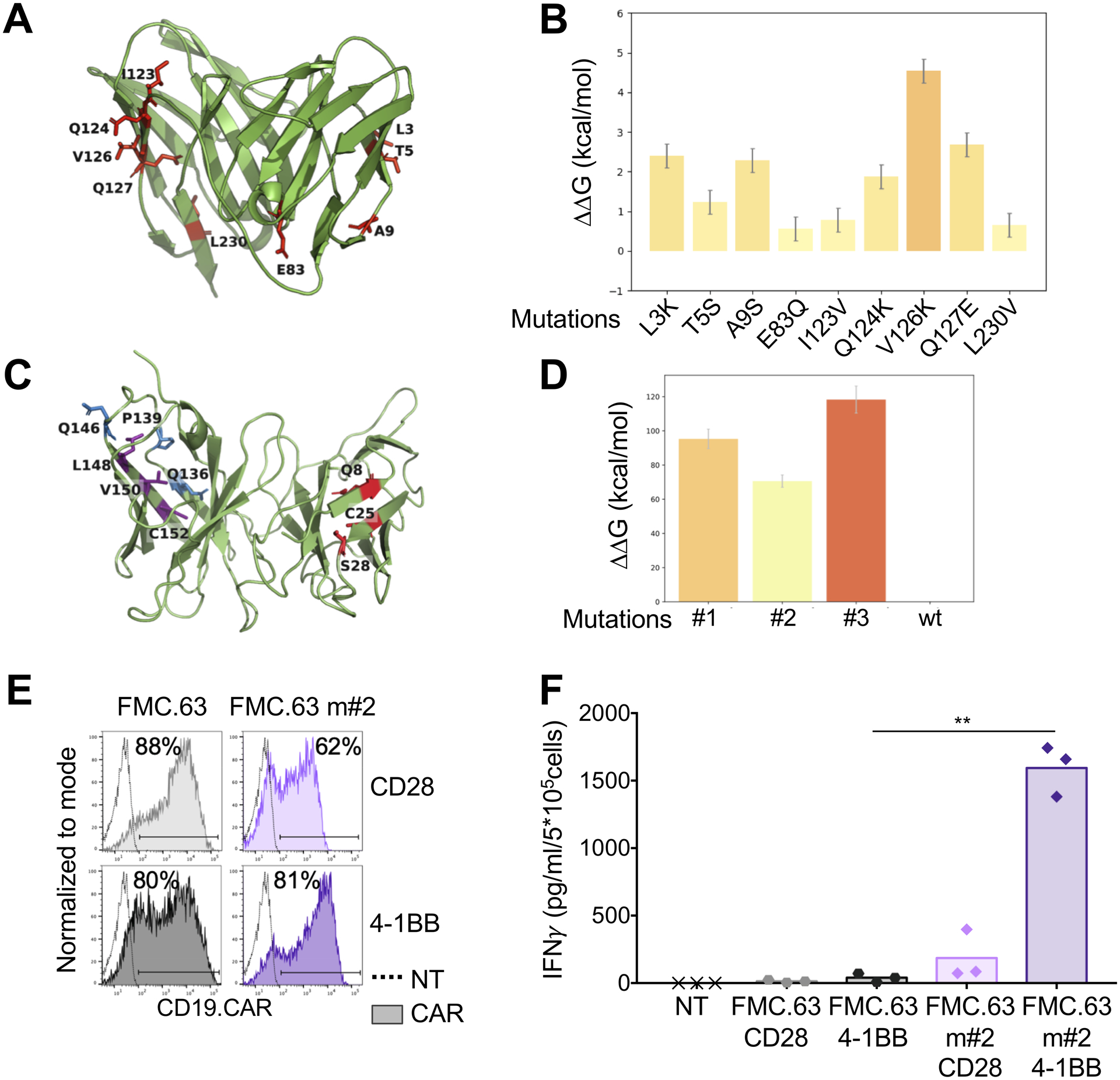
A. Structural conformation of the scFv 763.74(B) generated through computational modeling. Protein is shown in cartoon representation and FWR mutations in stick representation. B. Chosen amino acid mutations evaluated for their influence on scFv 763.74(B) stability (n=50, mean and SD shown). C. Structural conformation of the scFv FMC.63 generated through computational modeling. Protein is shown in cartoon representation and FWR mutations in stick representation. D. Chosen amino acid mutations (Q8W|C25R|S28P, Q136W|P139L|Q146P, and L148Y|V150W|C152W) evaluated for their influence on the scFv FMC.63 stability (n=50, mean and SD shown). E. Representative flow cytometry plots showing CAR expression in T cells engineered with the scFv FMC.63 CARs and the destabilized scFv FMC.63 m#2 CARs encoding either CD28 or 4–1BB endodomains. CAR expression was assessed using an anti-idyotipic antibody followed by the staining with a secondary rat anti-mouse antibody. F. Quantification of IFNγ released into supernatants by T cells expressing the different CARs without CAR-specific activation after 24 hours (n=3, mean shown). **P=0.0047, paired t test.
