Figure 1.
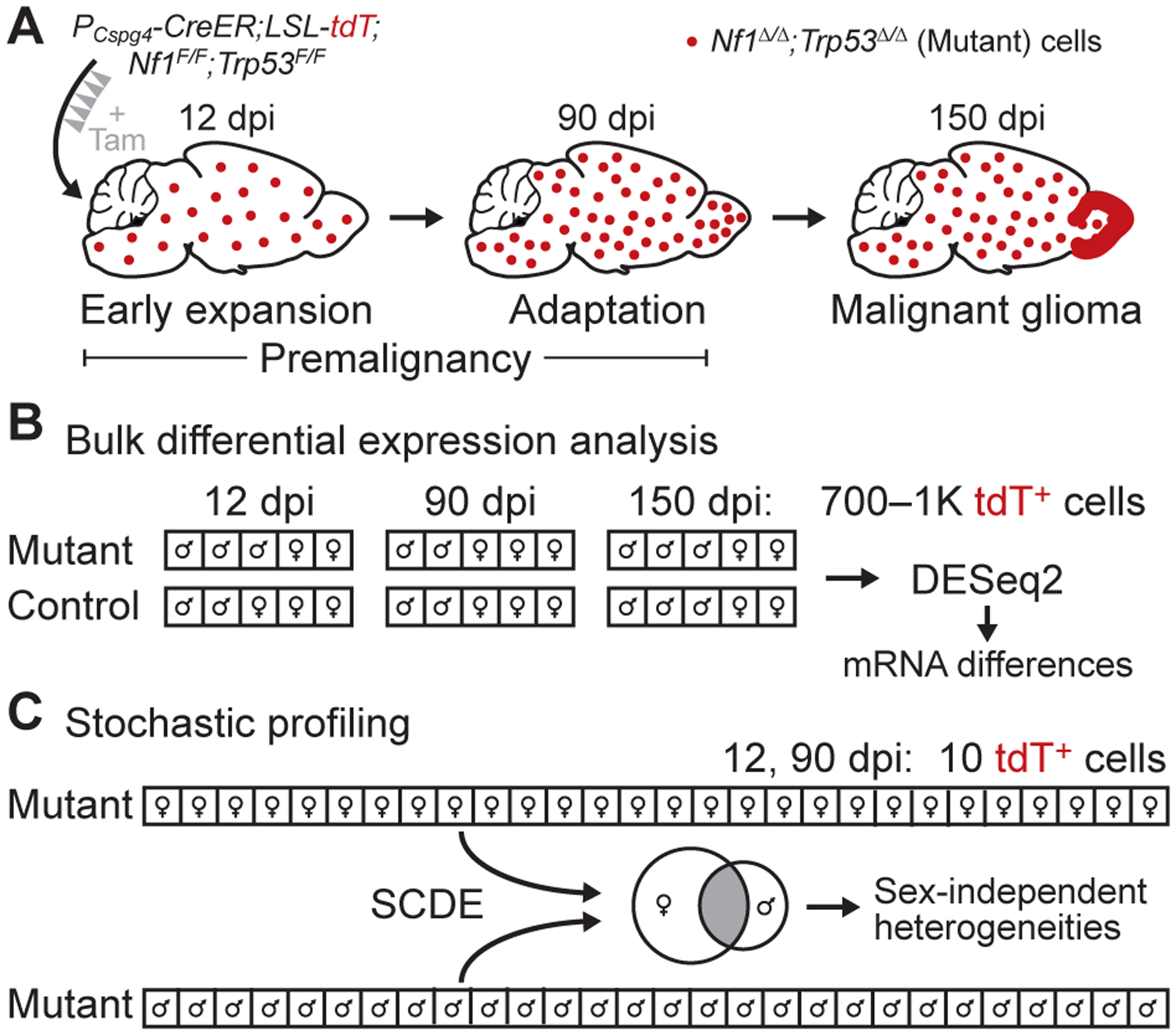
Transcriptomic analysis of bulk expression differences and regulatory heterogeneities in a GEMM of glioma. A, Summary of the glioma GEMM. Floxed alleles of Nf1 and Trp53 are inducibly deleted by tamoxifen (Tam) activation of CreER localized to oligodendrocyte precursor cells with a Cspg4 promoter (PCspg4). The resulting Nf1Δ/ΔTrp53Δ/Δ (mutant) cells are labeled with tdTomato (tdT) and expand in the days post-induction (dpi) to form gliomas preferentially in the olfactory bulb (15). B, Experimental plan for bulk differential expression analysis. Large numbers of labeled cells from mutant and control animals were collected by LCM at the indicated dpi, linearly amplified, and sequenced for differential expression (n = 4–7 animals at roughly equal male-female proportion for each group). C, Experimental plan for heterogeneity analysis by stochastic profiling (17,19). Many 10-cell pools of mutant cells (n = 28 of each sex) were collected, measured by 10cRNA-seq (18), and assessed for regulatory heterogeneity with the abundance-dependent dispersion module of the SCDE package (42,43). Male and female candidates were intersected to arrive at the final set of regulatory heterogeneities.
