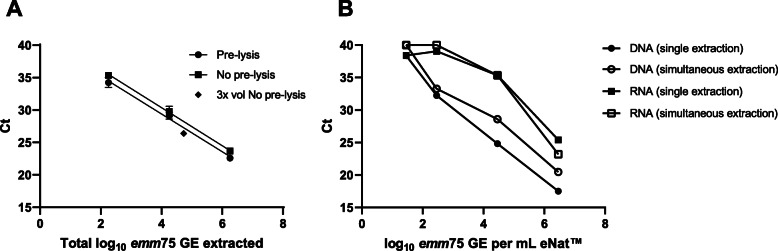
Fig. 2.
Optimisation of the method for isolation of DNA and RNA from throat swabs in eNat. a Pre-lysis versus No pre-lysis extractions using the combined RNA+DNA approach. eNats were spiked with S. pyogenes M75 at three concentrations and 200l aliquots (1.8102, 1.8104, 1.8106 total GE) underwent DNA extraction using a chemical Pre-lysis step () or No pre-lysis (). A larger 600l aliquot (3x vol) of eNat medium spiked with M75 (5.4104 total GE) was processed without pre-lysis (). Data shown in total GE to account for differences in input bacteria. Each point represents the mean and SD of two eNats tested in duplicate qPCRs. b eNats containing a range of M75 were used to test separate column extractions (RNA-only or DNA-only ) versus combined RNA+DNA isolation (RNA and DNA). The emm75 qPCR was used to assess DNA whilst RNA was converted to cDNA and tested in a SYBR Green qPCR for the housekeeping gene, gyrA. Each point represents the mean and SD of three eNats tested in duplicate qPCRs targeting the emm75 gene