FIGURE 2.
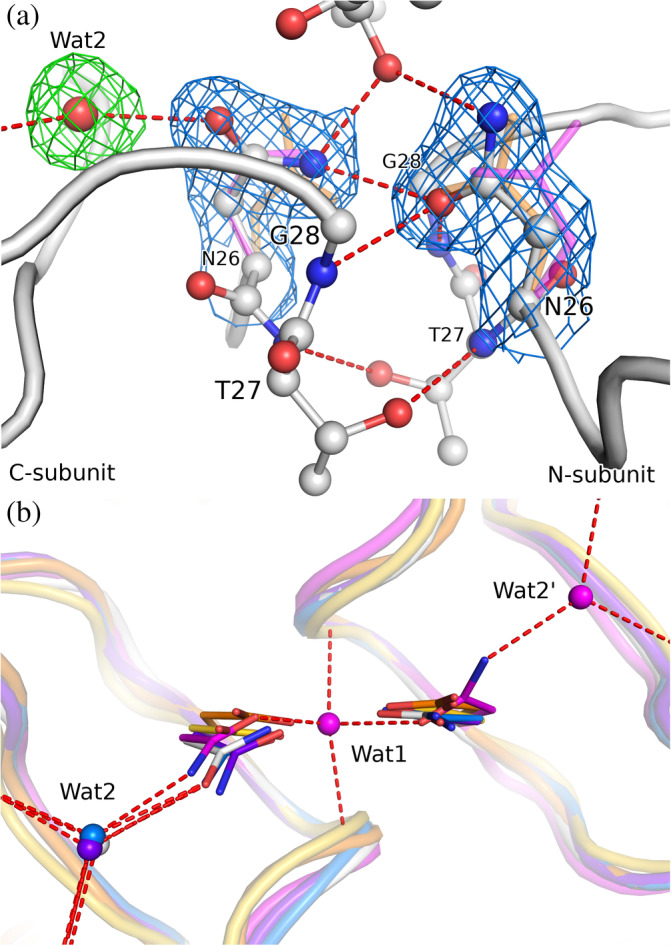
(a) The active site of 3M2 (chains A/B, white) with H‐bond network including a water molecule. The side chains of the Asn26 residues are shown in 2mF o–DF c electron density (blue) contoured at 1.0σ, the water molecule (Wat2) is shown in mF o‐DF c polder electron density (green) contoured at 3.0σ. For comparison, the Asp25 side chain of aligned HIV‐1 PR (PDB ID 4hvp, 19 orange) and Asn26 of a different (P21) crystal form of M‐PMV PR (6s1v, magenta) are shown. (b) A perpendicular view of the active site comparing the conformation of the catalytic Asp (or Asn mutation) residues, together with H‐bonded water sites. Wat1 is a water molecule buried between active site loops in the monoclinic structure 6s1v; Wat2 interacts with the main‐chain carbonyls and the catalytic Asp/Asn side chain. The superposition comprises 3M2 (white), M‐PMV PR crystallized in P21 (6s1v, magenta), HIV‐1 PR: holo (4hvp, orange) and apo form (3hvp, 20 yellow) plus two complexes with water molecules within H‐bond distance of the active site (5kr1, blue; 1n49, purple)
