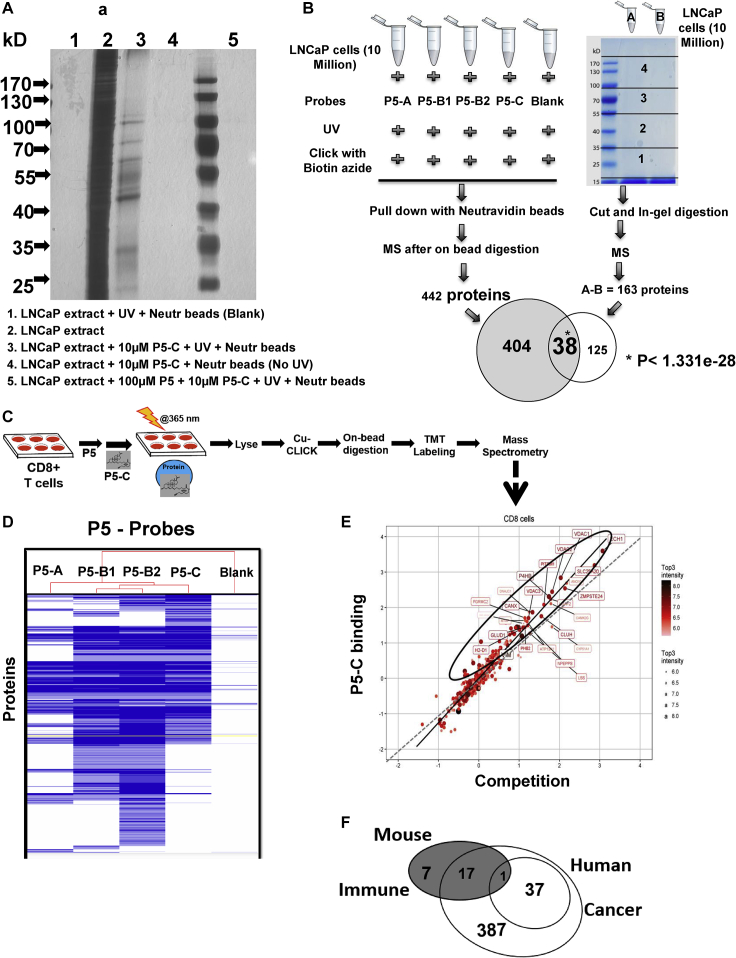
Figure 3.
P5 probes enable mass spectrometry profiling of P5-binding proteins in live cells
(A) Gel profiling and specificity of P5-binding proteins with P5-C in LNCaP cell extracts. About 400 μg of LNCaP protein in vitro was mixed either with P5-C alone or in the presence of 10X P5 (competition assay), exposed to UV, and loaded onto lanes 3 and 5, respectively, in a 10% SDS-PAGE. Two control experiments were run in parallel, one without P5-C to check background binding to the beads (lane 1) and another without UV to check the proper activation of the diazirine within the linker (lane 4). Lane 2 has 50 μg of total LNCaP proteins. Lanes 3, 4, and 5 were loaded with the final pulldown from 400 μg of LNCap extracts in laemmli buffer. The gel was stained with silver and imaged.
(B) Experimental design to reveal the identity of P5-interacting proteins in LNCaP cells using two approaches with 10 million live cells. In the right-hand panel gel image, “A” represents pulldown with10 μM P5-C and “B” represents pulldown after competition reaction (100 μM P5+10 μM P5-C). The p value calculations were done as per the hypergeometric probability formula from Chapter 6.1 of Numerical Recipes in C: The Art of Scientific Computing (ISBN 0-521-43108-5) (Press et al., 1992) as found in http://nemates.org/MA/progs/representation.stats.html.
(C) Experimental design to reveal the identity of P5-interacting proteins in live CD8+ T cells.
(D) Hierarchical clustering of all proteins pulled down using all four probes shows the structure-dependent bioactivity of the probes.
(E) Twenty-three P5-interacting proteins in CD8+ T cells. The proteins extracted with P5-C either alone or after competition with P5. P5-C binding proteins either in the presence of P5 (competition assay) or in its absence (experiment) were plotted on a log scale after subtracting the background. The regression equation fitting the two variables is represented by the solid straight line, whereas the dotted line represents the X = Y linear relation showing no variation. The dots in ellipses represent the P5-C binding proteins whose binding can be competed out in the presence of P5.
(F) Common and specific P5-interacting proteins in human prostate cancer and mouse immune cells. P5-interactomes from human prostate cancer (two concentric empty circles) and murine CD8+ T cells (filled circle) have 16 proteins in common. Among them, only one protein was common to CD8+ T cells and core “P5-binding proteins’ from LNCaP. Another 17 CD8+ T cell proteins were found in the 404 “potential P5-binding proteins” from LNCaP. Seven and four hundred twenty-four (37 + 387) proteins are specific to CD8+ T cells and LNCaP, respectively.