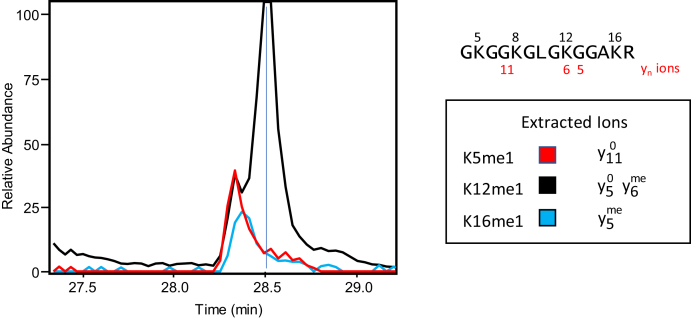
Fig. 6.
Monomethylation profile for histone H4 K5, K12, and K16 from Z-138 cancer cells. Extracted ion chromatograms from PRM analysis of m/z 726.9178, the [M+2H]2+ precursor ion for the tryptic peptide H4 (4–17) + me1. These ions confirm methylation on different lysine residues within the 4–17 peptide. The stoichiometry for K5me1 was calculated using the intensity of the unmethylated y11 ion (m/z 1154.6698), which is unique for K5me1. The stoichiometry for K16me1 was calculated using the intensity of the methylated y5 ion (m/z 544.3232), which is unique for K16me1. The remaining methylation was assigned to K12me1, which is identified by a combination of the unmethylated y5 ion (m/z 530.3056) and the methylated y6 ion (m/z 714.4268). No unequivocal evidence was found for methylation on K8. Labeled spectra are shown in supplemental Fig. S6.