Figure 1.
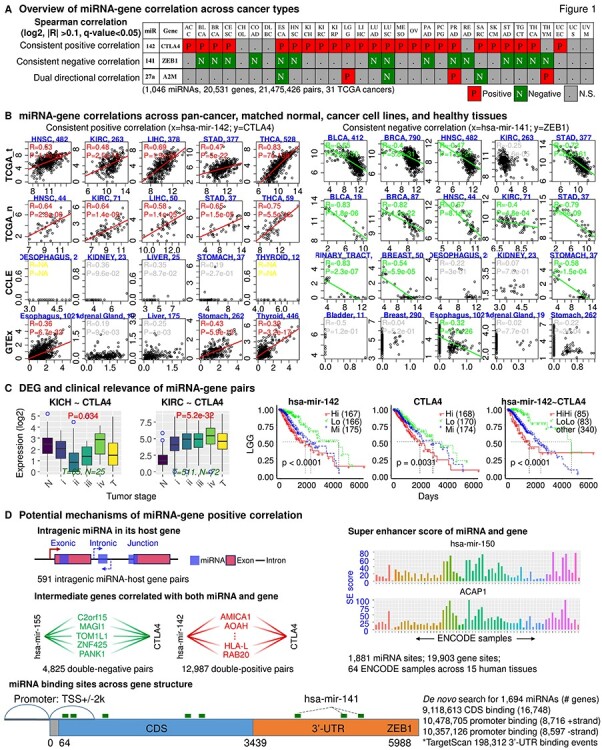
Overview of miRactDB. (A) General output upon miRNA–gene query: correlation profile across TCGA cancers. Representative results are shown for consistent positive and negative correlations, as well as correlation with dual direction in different cancers. N.S., not significant. (B) Example correlation patterns for miR-142~CTLA4 (left) and miR-141~ZEB1 (right) across representative cancer/tissue types in TCGA (both tumour and normal), CCLE (cancer cell line) and GTEx (non-disease tissue). (C) Left, gene expression along normal and different tumour stages; right, clinical relevance by survival analysis based on individual or combined miRNA–gene expression. (D) Evidence for five hypotheses regarding miRNA-associated/miRNA-directed gene activation as indicated in each panel: intragenic miRNA–host gene co-expression; double-negative and double-positive indirect regulation; co-regulation by same epigenetic factor based on ENCODE ChIP-seq for H3K27ac; and direct binding and upregulation by scanning potential miRNA binding sites on the coding and promoter regions of queried gene with TargetScan script.
