Table 3.
Average and standard deviation of precision, Recall and F1-score obtained by each tool in the Strand-Blind full transcript sets analyzed. Bold font highlight the higher value for each group.No results are available for ESTScan in fungal transcripts due to the absence of a fungal model
| Group | Predictor | Precision | Recall | F1-score |
|---|---|---|---|---|
| Vertebrates | CodAn |
0.99  0.00 0.00
|
0.98  0.01 0.01
|
0.99  0.01 0.01
|
| ESTscan | 0.36  0.02 0.02 |
0.36  0.02 0.02 |
0.88  0.13 0.13 |
|
| TransDecoder | 0.18  0.04 0.04 |
0.18  0.04 0.04 |
0.93  0.05 0.05 |
|
| Prodigal | 0.27 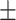 0.05 0.05 |
0.27 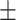 0.05 0.05 |
0.93  0.02 0.02 |
|
| GeneMarkS-T | 0.50  0.00 0.00 |
0.49  0.00 0.00 |
0.49  0.00 0.00 |
|
| Invertebrates | CodAn |
0.99  0.02 0.02
|
0.95  0.03 0.03
|
0.97  0.02 0.02
|
| ESTscan | 0.31  0.10 0.10 |
0.21  0.13 0.13 |
0.24  0.12 0.12 |
|
| TransDecoder | 0.29  0.08 0.08 |
0.28  0.07 0.07 |
0.29  0.08 0.08 |
|
| Prodigal | 0.39  0.05 0.05 |
0.39  0.05 0.05 |
0.39  0.05 0.05 |
|
| GeneMarkS-T | 0.49 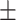 0.01 0.01 |
0.48  0.01 0.01 |
0.48 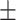 0.01 0.01 |
|
| Plants | CodAn |
0.99  0.01 0.01
|
0.96 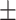 0.04 0.04
|
0.98  0.02 0.02
|
| ESTscan | 0.35  0.06 0.06 |
0.35  0.06 0.06 |
0.35  0.06 0.06 |
|
| TransDecoder | 0.22 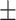 0.13 0.13 |
0.21  0.12 0.12 |
0.21  0.12 0.12 |
|
| Prodigal | 0.35  0.05 0.05 |
0.35 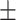 0.05 0.05 |
0.35 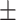 0.05 0.05 |
|
| GeneMarkS-T | 0.49  0.01 0.01 |
0.48 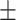 0.02 0.02 |
0.49 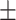 0.01 0.01 |
|
| Fungi | CodAn |
0.98 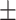 0.01 0.01
|
0.94 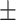 0.04 0.04
|
0.96  0.02 0.02
|
| ESTscan | NA | NA | NA | |
| TransDecoder | 0.21  0.11 0.11 |
0.20  0.10 0.10 |
0.21 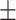 0.11 0.11 |
|
| Prodigal | 0.36  0.06 0.06 |
0.35  0.06 0.06 |
0.36  0.06 0.06 |
|
| GeneMarkS-T | 0.49  0.01 0.01 |
0.47  0.02 0.02 |
0.48 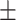 0.01 0.01 |
