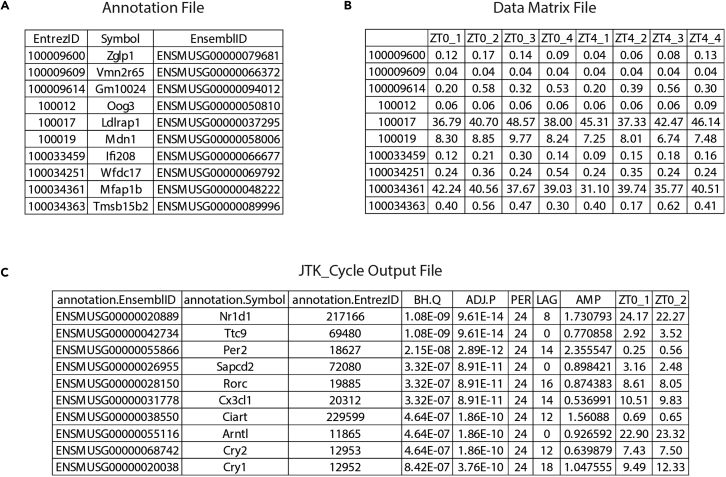
Figure 4.
Input and output files of JTK_CYCLE
(A) Correctly structured "annotation file", as required to run JTK_CYCLE. Only the first 10 rows of data are shown, along with the appropriate column headers. In this example, EntrezID, HUGO symbol (Symbol), and Ensembl ID gene annotations are included.
(B) A correctly structured "data matrix file", as required to run JTK_CYCLE. Only the first 10 rows and 9 columns of data are shown, along with the appropriate column headers. The first column lists the EntrezIDs of all genes with expression quantified by RNA-seq. Subsequent columns indicate the RPKM expression values of each gene at each time-point, in chronological order (ZT0, ZT4, ZT8…). Replicates are placed in adjacent columns (ZT0_1, ZT0_2…).
(C) A "JTK_CYCLE Output File" generated from the example input files in (A) and (B), and sorted by the statistical significance of circadian gene expression. The first three columns list the different gene annotations present in the "annotation file", and the following five columns give the circadian parameters of each gene as calculated by JTK_CYCLE. In the final columns, the RPKM values used by the algorithm are repeated. The first 10 rows and 10 columns of data are shown, along with the column headers. In this example, JTK_CYCLE identified 2035 rhythmic transcripts (ADJ.P <0.01) in WT liver, which represented 11.25% of the total measured transcripts. BH.Q, Benjamini–Hochberg q-value; ADJ.P, adjusted p-value; PER, period; LAG, phase; AMP, amplitude.