Figure 1:
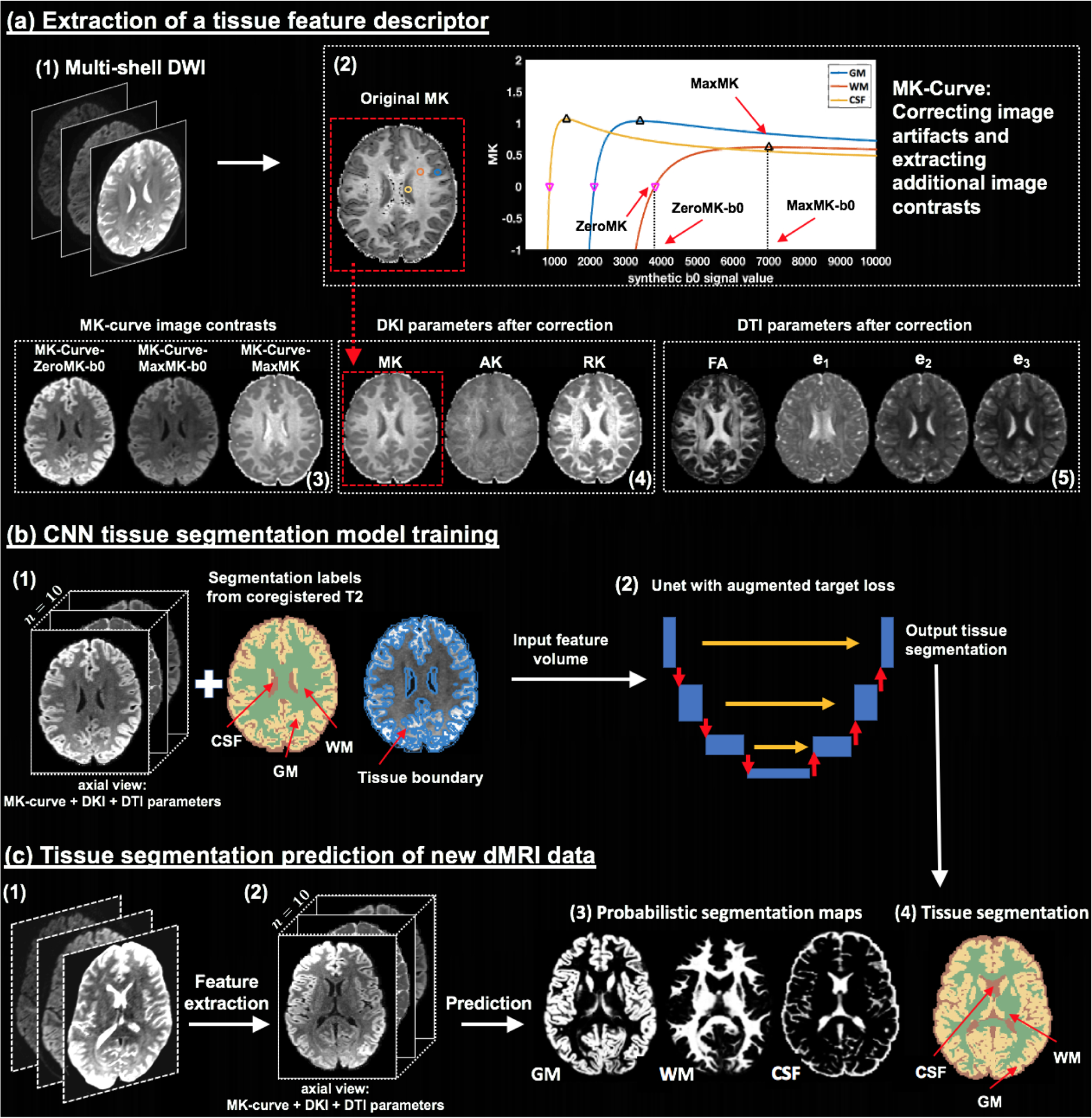
Method overview. Given input dMRI data (a1), an MK-Curve is computed for each voxel (a2, showing three example voxels). The MK-Curve is used to correct implausible DKI and DTI parameters, and to derive three additional image contrasts that are useful to identify tissue types: ZeroMK-b0, MaxMK-b0 and MaxMK (a3). A total of 10 features are computed: the 3 MK-Curve contrasts (a3), 3 DKI maps (a4) and 4 DTI maps (a5). The corrected images no longer have implausible values (e.g., original MK map (a2) versus corrected MK map (a4); in a red frame). The 3D volumes of the 10 features along with the segmentation labels and the tissue boundary segmentation computed from co-registered T2w data (b1) are used to train a CNN with a Unet architecture and with a novel augmented target loss function (b2). For new dMRI data (c1), the trained CNN is applied on the computed 3D volume of the 10 features in the dMRI space directly (c2), not requiring inter-modality registration. The final prediction output includes a probabilistic segmentation map for each tissue type (c3) and a segmentation label per voxel (c4).
