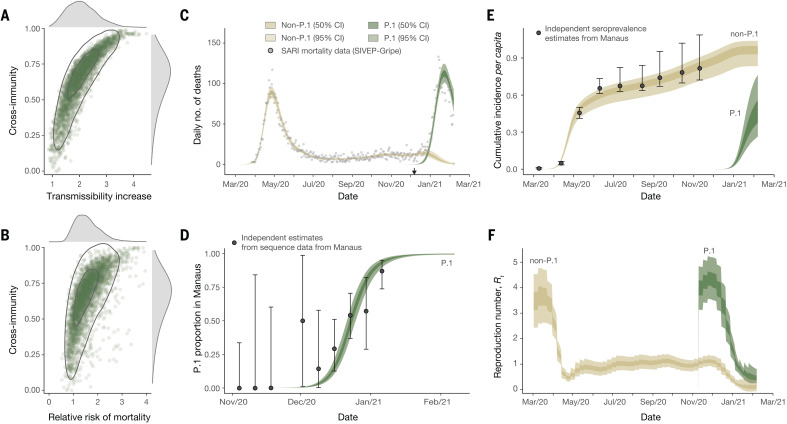
Fig. 4. Estimates of the epidemiological characteristics of P.1 inferred from a multicategory Bayesian transmission model fitted to data from Manaus, Brazil.
(A) Joint posterior distribution of the cross-immunity and transmissibility increase inferred through fitting the model to mortality and genomic data. Gray contours indicate posterior density intervals ranging from the 95 and 50% isoclines. Marginal posterior distributions for each parameter shown along each axis. (B) As for (A), but showing the joint-posterior distribution of cross-immunity and the inferred relative risk of mortality in the period after emergence of P.1 compared with the period prior. (C) Daily incidence of COVID-19 mortality. Points indicate severe acute respiratory mortality records from the SIVEP-Gripe database (67, 69). Brown and green ribbons indicate model fit for COVID-19 mortality incidence, disaggregated by mortality attributable to non-P.1 lineages (brown) and the P.1 lineage (green). (D) Estimate of the proportion of P.1 infections through time in Manaus. Black data points with error bars are the empirical proportion observed in genomically sequenced cases (Fig. 3A), and green ribbons (dark = 50% BCI, light = 95% BCI) are the model fit to the data. (E) Estimated cumulative infection incidence for the P.1 and non-P.1 categories. Black data points with error bars are reversion-corrected estimates of seroprevalence from blood donors in Manaus (2). Colored ribbons are the model predictions of cumulative infection incidence for non-P.1 lineages (brown) and P.1 lineages (green). These points are shown for reference only and were not used to fit the model. (F) Bayesian posterior estimates of trends in reproduction number Rt for the P.1 and non-P.1 categories.