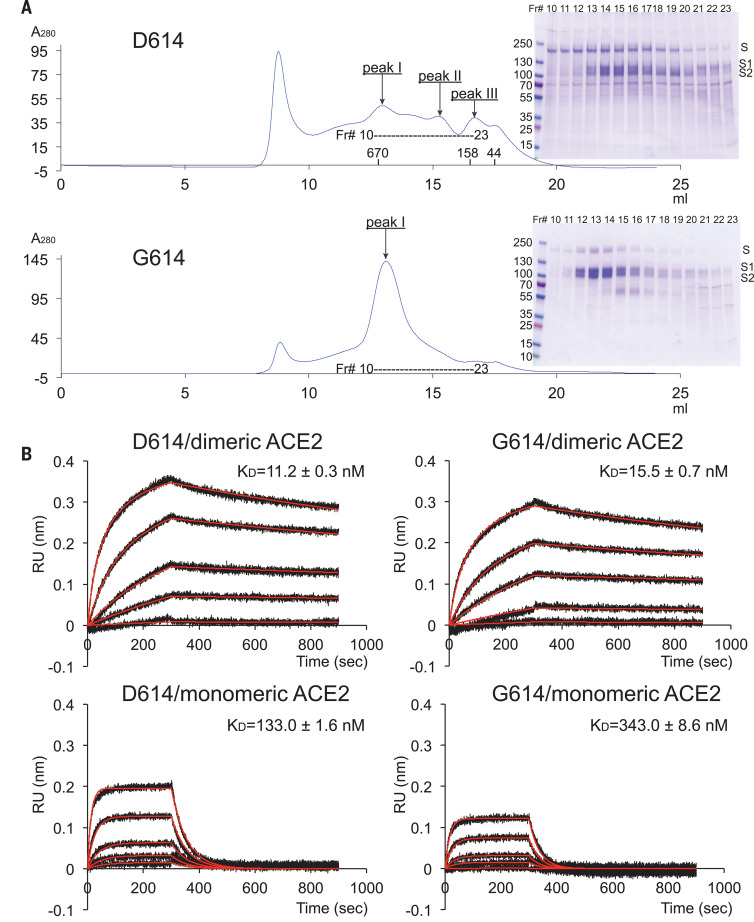
Fig. 1. Characterization of the purified full-length SARS-CoV-2 S proteins.
(A) The full-length SARS-CoV-2 S protein carrying either D614 or G614 was extracted and purified in detergent n-dodecyl--d-maltopyranoside (DDM) and further resolved by gel-filtration chromatography on a Superose 6 column. The molecular weight standards include thyoglobulin (670 kDa), -globulin (158 kDa), and ovalbumin (44 kDa). Peak I is the prefusion S trimer; peak II is the postfusion S2 trimer, and peak III is the dissociated monomeric S1. The insets show peak fractions that were analyzed by Coomassie-stained SDS-PAGE. Labeled bands are S, S1, and S2. Fr#, fraction number. (B) Binding analysis of fractions of peak I in (A) with soluble ACE2 constructs by BLI. The purified S proteins were immobilized to AR2G biosensors and dipped into the wells containing ACE2 at various concentrations (5.56 to 450 nM for monomeric ACE2; 2.78 to 225 nM for dimeric ACE2). Binding kinetics was evaluated using a 1:1 Langmuir binding model for the monomeric ACE2 and a bivalent model for dimeric ACE2. The sensorgrams are in black and the fits in red. Binding constants are also summarized here and in table S1. All experiments were repeated at least twice with essentially identical results. KD, dissociation constant (binding affinity); RU, response unit.