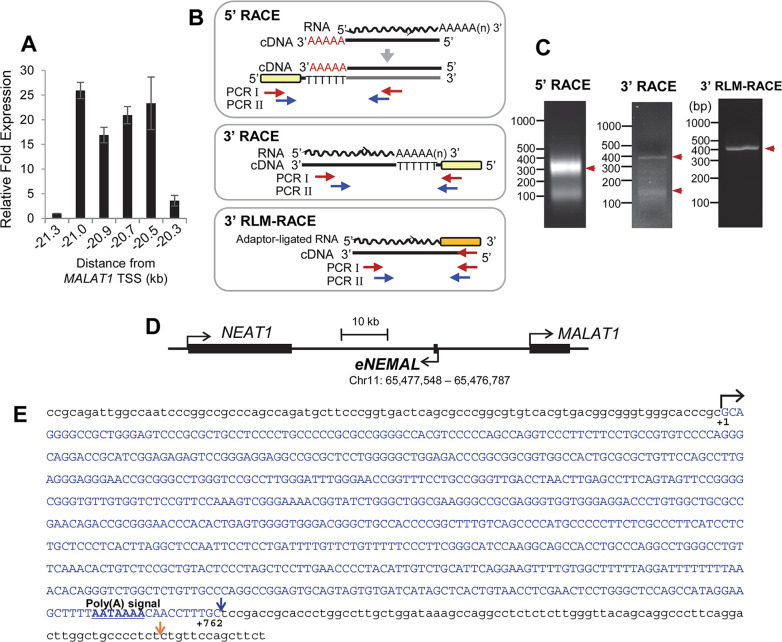
Fig 2. eNEMAL is a polyadenylated and unspliced eRNA transcribed from the MALAT1 -20kb enhancer.
(A) RT-qPCR tiling assay for MCF7 endogenous eRNA expression with coordinates relative to the MALAT1 transcription start site. Data shown as mean ± SD, n = 3. (B) Schematics of 5’ RACE, 3’ RACE, and RNA ligase-mediated 3’ RACE (3’ RLM-RACE), and the locations of the primers used. RNA used for RACE was isolated from MCF7 cells after 24 hours hypoxic exposure. Red arrows, primer sets used for the first PCR; Blue arrows, primer sets used for the second PCR; Yellow bar, adaptor; Orange bar, oligonucleotide ligated to RNA. In 5’RACE, AAAAA in red font indicates the DNA sequences added by the terminal transferase reaction. (C) Gel images of 5’ RACE, 3’ RACE and 3’ RLM-RACE products. Bands indicated with red arrowheads were cut and sequenced. (D) Genomic location of the identified eRNA, eNEMAL, relative to the MALAT1 and NEAT1 gene bodies. The position of the eNEMAL gene was determined using hg38 as reference genome. (E) Full sequence of the eNEMAL gene. Blue arrow downstream of the poly(A) signal indicates the poly(A)-tailed 3’ end of the eNEMAL transcript determined by 3’ RACE. Orange arrow indicates a non-poly(A)-tailed alternative 3’ end determined by 3’ RLM-RACE.