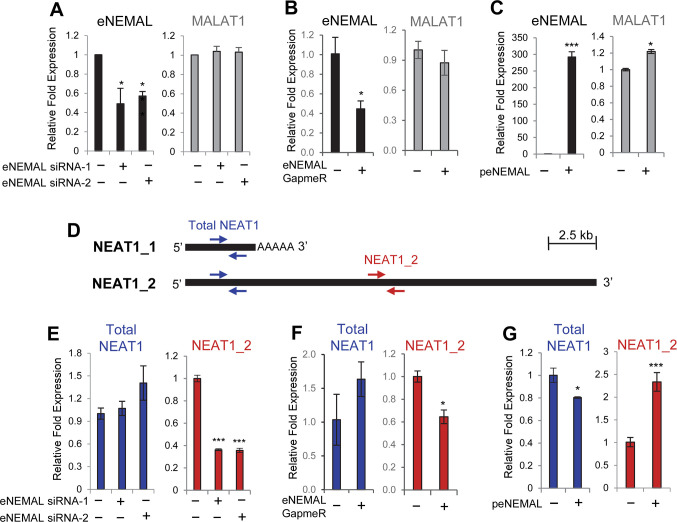
Fig 5. eNEMAL regulates NEAT1 isoform switching, resulting in upregulation of the NEAT1_2 long isoform in MCF7 breast cancer cells.
(A, B and C) RT-qPCR measuring MALAT1 expression after knocking down or overexpressing eNEMAL. MCF7 cells were transfected with two different siRNAs targeting eNEMAL and maintained under hypoxia (A), transfected with GapmeR targeting eNEMAL and maintained under hypoxia (B), or transfected with eNEMAL-overexpressing plasmid (pCDH-eNEMAL) and maintained under normoxia (C). RNA was isolated 48 hours after siRNA/GapmeR/plasmid transfection and used for RT-qPCR. (D) qPCR primer scheme to detect total NEAT1 (both the short isoform NEAT1_1 and the long isoform NEAT1_2; blue arrows) and NEAT1_2 only (red arrows). (E, F and G) RT-qPCR measuring total NEAT1 and NEAT1_2. MCF7 cells were transfected with eNEMAL siRNA and maintained under hypoxia (E), transfected with GapmeR targeting eNEMAL and maintained under hypoxia (F), or transfected with eNEMAL-overexpressing plasmid (pCDH-eNEMAL) and maintained under normoxia (G) prior to RNA isolation as described above. Data shown as mean ± SD, n = 4, * P < 0.01, *** P < 0.001.