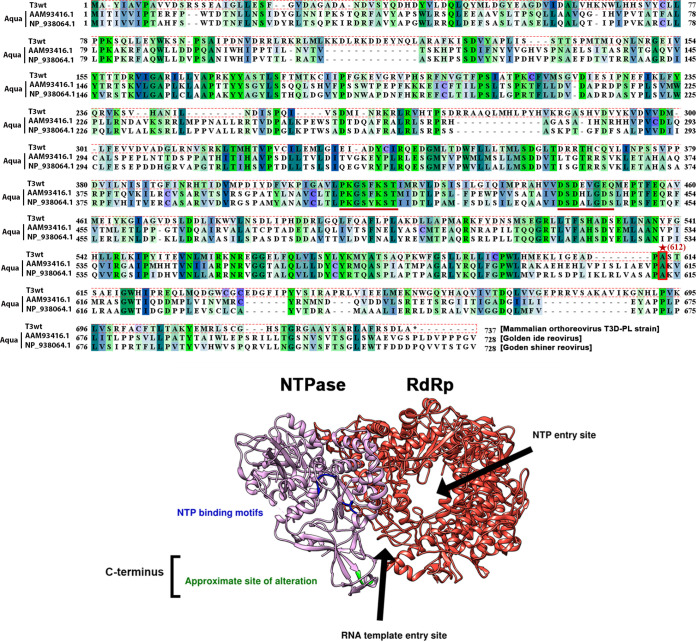
FIG 10.
The T3v10M1 μ2 amino acid replacement is located in the uncharacterized C terminus. (Top) Sequences of T3wt μ2 (top sequence), were compared with those of two subspecies of Aquareovirus homolog VP5. The structure of VP5 has been recently published (bottom) from the middle sequence. The NTPase motifs are well conserved (underlined), but the T3wt residue 612 alanine (site with an *, replaced by valine in T3v10M1) is located in the C terminus, which is not conserved among the three viruses. Note that all of the sequences were downshifted when the multiple alignment was performed. (Bottom) A 3D model of the transcription complex composed of the polymerase VP2(λ3) and its cofactor VP5(μ2). Due to the high homology between VP5 and μ2, a small region of the C terminus has been highlighted in green to represent the T3wt μ2 A612V alteration, which is located at the entrance of the RNA template entry site but not in close proximity to the NTP binding motifs. The C terminus of μ2 has not been characterized previously and is not associated with any known function.