Human coronaviruses (HCoV) are recognized respiratory pathogens. The emergence of the novel pathogenic member of this family in December 2019 (SARS-CoV-2, which causes COVID-19) poses a global health emergency.
KEYWORDS: coronavirus, SARS-CoV-2, CNS, neuroinvasion, neuropropagation, olfactory sensory neuron, S protein, cleavage, type 1 interferon, IFN-1, central nervous system infections
ABSTRACT
Human coronaviruses (HCoV) are respiratory pathogens which have been known since the 1960s. In December 2019, a new betacoronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), was reported, and it is responsible for one of the biggest pandemics of the last 2 centuries. Available evidence suggests that similar to the case with the HCoV-OC43 strain, SARS-CoV-2 neuroinvasion is associated with potential neurological disorders. Coronavirus infection of the central nervous system (CNS) is largely controlled by a viral factor, the spike glycoprotein (S), and a host factor, innate immunity. However, the interaction between these two factors remains elusive. Proteolytic cleavage of the S protein can occur at the interface between receptor binding (S1) and fusion (S2) domains (S1/S2), as well as in a position adjacent to a fusion peptide within S2 (S2′). In this study, using HCoV-OC43 as a surrogate for SARS-CoV-2, we determined that both S protein sites are involved in neurovirulence and are required for optimal CNS infection. Whereas efficient cleavage at S1/S2 is associated with decreased virulence, the potentially cleavable putative S2′ site is essential for efficient viral infection. Furthermore, type 1 interferon (IFN-1)-related innate immunity also plays an important role in the control of viral spread toward the spinal cord, by preventing infection of ependymal cells. Our results underline the link between the differential S cleavage and IFN-1 in the prevention of viral spread, to control the severity of infection and pathology in both immunocompetent and immunodeficient mice. Taken together, these results point toward two potential therapeutic antiviral targets: cleavage of the S protein in conjunction with efficient IFN-1-related innate immunity to prevent or at least reduce neuroinvasion, neural spread, and potential associated neurovirulence of human coronaviruses.
IMPORTANCE Human coronaviruses (HCoV) are recognized respiratory pathogens. The emergence of the novel pathogenic member of this family in December 2019 (SARS-CoV-2, which causes COVID-19) poses a global health emergency. As with other coronaviruses reported previously, invasion of the human central nervous system (CNS), associated with diverse neurological disorders, was suggested for SARS-CoV-2. Herein, using the related HCoV-OC43 strain, we show that the viral spike protein constitutes a major neurovirulence factor and that type 1 interferon (IFN-1), in conjunction with cleavage of S protein by host proteases, represents an important host factor that participates in the control of CNS infection. To our knowledge, this is the first demonstration of a direct link between cleavage of the S protein, innate immunity, and neurovirulence. Understanding mechanisms of viral infection and spread in neuronal cells is essential to better design therapeutic strategies, and to prevent infection by human coronaviruses such as SARS-CoV-2 in the human CNS, especially in vulnerable populations such as the elderly and immunocompromised individuals.
INTRODUCTION
Human coronaviruses (HCoV) were first identified in the 1960s (1–3). Since the beginning of the 21st century, several new members of the Coronaviridae family have been identified, including five that infect humans: severe acute respiratory syndrome coronavirus (SARS-CoV), HCoV-NL63, HCoV-HKU1, Middle East respiratory syndrome coronavirus (MERS-CoV), and, in December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiologic agent responsible for coronavirus disease 19 (COVID-19), associated with one of the largest pandemics of the last 2 centuries (4–12).
These viruses are recognized respiratory pathogens (1, 13, 14); however, SARS-CoV, SARS-CoV-2, HCoV-OC43, and HCoV-229E all have been shown to be naturally neuroinvasive and neurotropic in humans (14–18), and evidence has accumulated to support the idea that HCoV, including SARS-CoV-2, the COVID-19-inducing agent, could be associated with pathologies outside the respiratory tract, including the human central and peripheral nervous systems (19–37). Moreover, by infecting the neurons, SARS-CoV and HCoV-OC43 induce neuronal cell death associated or not with encephalitis in susceptible mice (38–41), and HCoV-OC43 and HKU-1, like SARS-CoV-2, have been associated with cases of encephalitis (42–45), acute disseminated encephalomyelitis (20), and encephalopathy (32) in humans.
Coronavirus infection is largely controlled by the spike glycoprotein (S), which bears both receptor binding and membrane fusion capabilities (46, 47). Being a class 1 viral fusion protein, this protein is proteolytically processed during infection of the host cell, a mechanism involved in the initiation of infection, tissue tropism, propagation and in eventual pathogenesis, including within the central nervous system (CNS) for several coronaviruses, such as SARS-CoV-2 (48–59). Different host proteases can cleave and activate the coronavirus spike, making these viruses very efficient at adapting to new environments, based on protease availability and on a multitude of mechanisms and strategies that have evolved to ensure infection (51, 57, 60). Proteolytic cleavage events that lead to membrane fusion can occur both at the interface between the receptor binding (S1) and fusion (S2) domains (S1/S2) and in a position adjacent to a fusion peptide within S2 (S2′) (61, 62). Fusion activation can be triggered by low pH, receptor binding, or a combination of the two, as the cleavage event occurs in the vicinity of the viral fusion peptide (63). We have reported that persistent HCoV-OC43 infections of human neural cell lines led to the appearance of point mutations in the S gene (64) and that these mutations were sufficient to significantly increase neurovirulence and modify neuropathology in mice (38). Moreover, a predominant mutation identified in the S gene of HCoV-OC43 variants detected in clinical isolates (Gly758Arg) introduces a furin-like protease cleavage site, RRSR↓R, in the S protein and modulates neurovirulence in mice (55). Type 1 interferon (IFN-1)-related innate immunity has also been shown to control coronavirus infection of the CNS (65, 66), and the host response to viral infection associated with this cytokine may involve host proteases in the airway and in the CNS (67–69).
While cleavage of the S protein has previously been shown to be important for infection in different cell culture models (56, 57, 70) and suggested to play a role in pathogenesis for the feline and murine coronaviruses as well as for SARS-CoV-2 (71–73), the contribution to neurovirulence is unclear. Here, we provide evidence of direct implication of putative cleavage sites (S1/S2 and S2′) in HCoV-OC43 neurovirulence and identify processes that may directly govern the severity and/or nature of human coronavirus neuropathogenesis. Our study also revealed that the IFN-1-related response plays a role in cell tropism and eventual dissemination within the CNS. Taken together, our results suggest that cellular proteases that modulate cleavage of the S protein, in combination with IFN-1, could be targeted to prevent CNS infection and potential neurovirulence of coronaviruses in humans.
RESULTS
Mutation at putative S protein cleavage sites influences HCoV-OC43 infectivity.
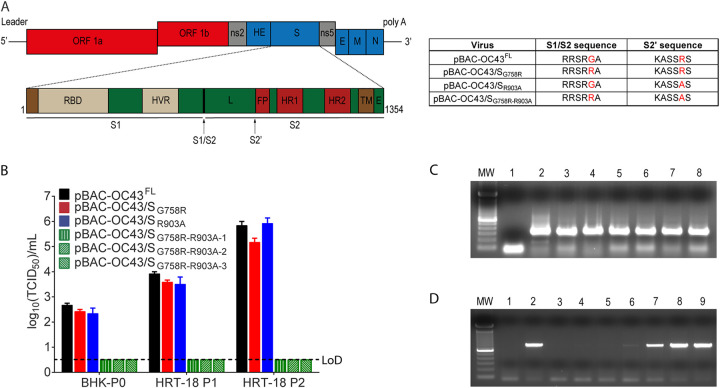
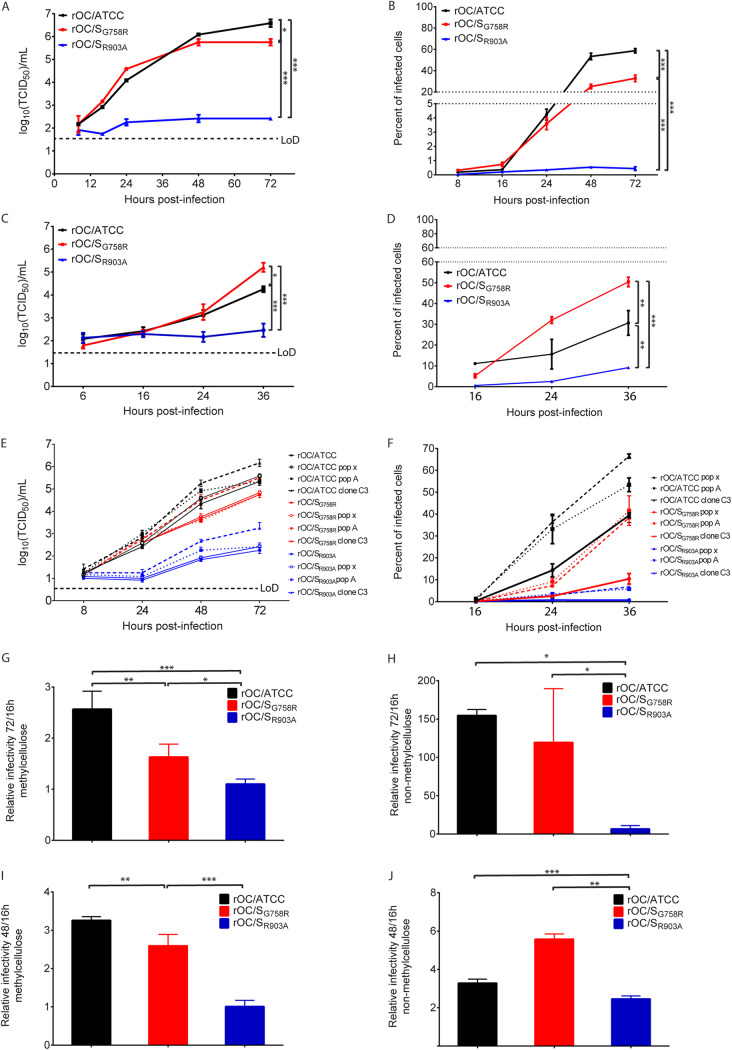
Coronaviruses have their S protein cleaved by host cell proteases during infection (51, 54–57, 61, 74, 75). We previously reported that cleavage of the HCoV-OC43 S protein at the S1/S2 site was not essential for viral infectivity, as HCoV-OC43 variants were able to infect cells despite the degree of cleavage at S1/S2. On the other hand, this cleavage was an important determinant of virus spreading in the CNS. By homology to other CoVs, we previously suggested (55) that a second cleavage site (S2′), already predicted by others through bioinformatics analysis and modeling (51), was present in the HCoV-OC43 S protein (Fig. 1A), and we have evaluated its importance during infection (51, 61, 74–76). We thus sought to investigate the potential relevance of the viral sequence KASSRS, located in the HCoV-OC43 S protein at the putative S2′ cleavage site before the peptide fusion (amino acids 899 to 904). Making use of our cDNA infectious clone (pBAC-OC43FL) (77), we wished to push further our understanding of the S cleavage mechanism and to evaluate the relative involvement of the identified S1/S2 and putative S2′ cleavage sites (Fig. 1A) during infection. Using recombineering (78), we generated cDNA infectious clones with mutations at either putative cleavage site (S1/S2 [pBAC-OC/SG758R] and S2′ [pBAC-OC/SR903A]) or at both sites (S1/S2 and S2′ [pBAC-OC/SG758R-R903A]) (Fig. 1A). Transfection of BHK-21 cells was performed, followed by amplification of infectious viruses in HRT-18 cells (77). However, using 3 different cDNA molecular clones for the double mutant (pBAC-OC/SG758R-R903A) (for which the entire S gene was sequenced), we were never able to harvest any detectable infectious viral particles compared to reference and single mutant viruses (Fig. 1B). This suggests that the S1/S2 and putative S2′ cleavage sites both play a role in the capacity to produce infectious virus. As expected, reverse transcription-PCR (RT-PCR) analysis revealed that transfections of all infectious clones produced viral RNA at comparable levels in BHK-21 cells (Fig. 1C). However, the viral RNA was already lost for the double mutant after the first viral amplification in HRT-18 cells (HRT-P1) (Fig. 1D), likely explaining the incapacity to detect infectious virus even after two rounds of amplification (HRT-P2) (Fig. 1B).
FIG 1.
Mutation at putative S protein cleavage sites influences the production of HCoV-OC43 infectious virus. (A) Representation of the full-length HCoV-OC43 genome found within the pBAC-OC43FL infectious clone with a schematic representation of the HCoV-OC43 S gene with the two cleavage sites: S1/S2 and putative S2′ cleavage site (black arrows). The S protein is composed of two subunits, S1, containing the receptor binding domain (RBD) and hypervariable region (HVR), and S2, containing the fusion subunit. L, linker region between S1/S2 and S2′ sites; FP, putative fusion peptide; HR1, heptad repeat 1; HR2, heptad repeat 2; TM, transmembrane domain; E, endodomain; ORF, open reading frame. Not drawn to scale. Data in table represent corresponding cleavage sequences of the different viruses, including reference pBAC-OC43FL. (B) Infectious virus production evaluated after transfection of BHK-21 cells (BHK-P0) and two rounds of amplification in HRT-18 cells (HRT-18 P1 and P2). (C) RT-PCR analysis for the presence of viral RNA after transfection of BHK21. Lane 1 is the negative control (ctrl) (transfected with wild-type pBAC-OC43FL without reverse transcription step), lane 2 is wild-type pBAC-OC43FL, lane 3 is pBAC-OC43/SG758R, lane 4 is clone 1 of pBAC-OC43/SR903A, lane 5 is clone 2 of pBAC-OC43/SR903A, lane 6 is clone 1 of pBAC-OC43/SG758R-R903A, lane 7 is clone 2 of pBAC-OC43/SG758R-R903A, and lane 8 is clone 3 of pBAC-OC43/SG758R-R903A. MW, molecular weight standards. (D) RT-PCR analysis for the presence of viral RNA in HRT-18 cells. Lane 1 is the negative ctrl (RNA extracted from OC43-infected cells without reverse transcription step), and lane 2 is the positive ctrl (RNA extracted from OC43-infected cells). Other lanes show RT-PCR performed on RNA extracted after two rounds of amplification (infection) in HRT-18 cells (HRT-18 P2): lane 3 comes from clone 1 of pBAC-OC43/SG758R-R903A, lane 4 comes from clone 2 of pBAC-OC43/SG758R-R903A, lane 5 comes from clone 3 of pBAC-OC43/SG758R-R903A, lane 6 comes from clone 1 of pBAC-OC43/SR903A, lane 7 comes from clone 2 of pBAC-OC43/SR903A, lane 8 comes from pBAC-OC43/SG758R, and lane 9 comes from wild-type pBAC-OC43FL.
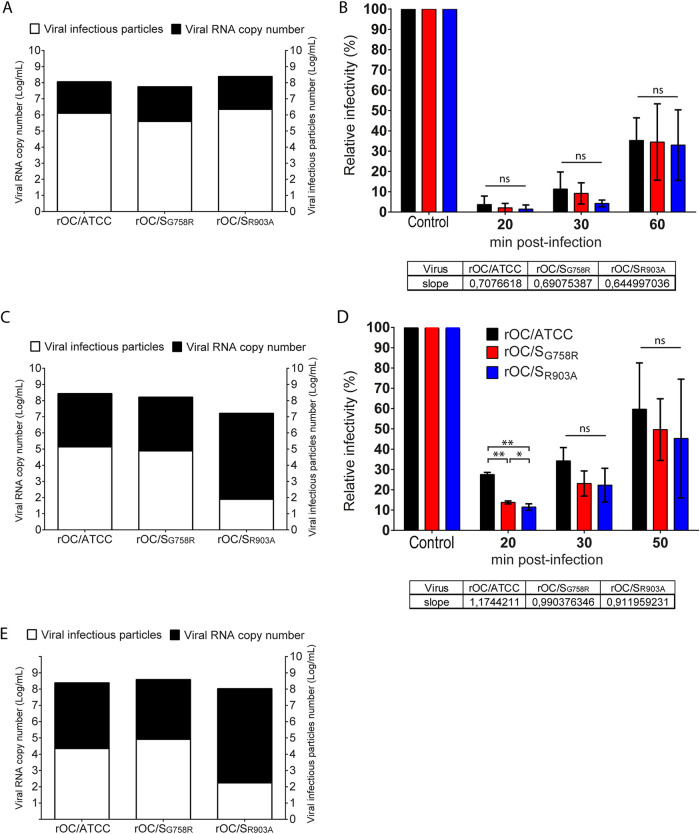
Given that no difference in infectivity between the rOC/SG758R virus and reference rOC/ATCC virus was observed in our previous study (55), we next sought to evaluate if inactivation of the S2′ putative cleavage site would impact infection. Statistical analysis revealed no significant difference in infectivity, defined as the ratio of infectious particles over total particles evaluated by the amount of viral RNA genome (55), between the three variants in viral stocks produced in human epithelial HRT-18 cells (average of 1 infectious viral particle for each 100 copies of viral RNA) and in the capacity to enter these epithelial cells (Fig. 2A and B). On the other hand, even though the infectivities were similar for rOC/ATCC and rOC/SG758R in neuronal cells, with an average of 1 infectious viral particle for each 1,000 and 3,000 viral RNA copies in LA-N-5 cells and primary cultures of the CNS, respectively, this parameter was drastically reduced for the rOC/SR903A variant. Indeed, the latter presented an average of infectivity of only 1 infectious viral particle for each 100,000 and 1,000,000 viral RNA copies in LA-N-5 neuronal cells and primary cultures of the CNS, respectively (Fig. 2C and E). The multiplicity of infection (MOI; calculated from titration on HRT-18 cells as described in Materials and Methods) used for infection of the different cell types was the same for all three variants in all experiments. Our data indicate that significantly fewer rOC/SG758R and rOC/SR903A virions were able to enter LA-N-5 neuronal cells in the early steps of infection. Indeed, after 20 min at 37°C, averages of only 11% and 14% for rOC/SR903A and rOC/SG758R viruses, respectively, entered neuronal cells, compared to 27% for rOC/ATCC. However, when evaluating global entry by calculating the slope over a 50-min time span, we determined that although a higher fraction of rOC/ATCC had entered cells at an earlier time point, overall all three variants were similarly successful at entering host cells (Fig. 2D).
FIG 2.
Mutation in S1/S2 and S2′ sites significantly impairs viral infectivity and entry of HCoV-OC43 into neuronal cells. An infectivity assay between viruses was conducted for quantification of viral RNA in viral stock (absolute quantity in RNA copy representative of total viral particles) over the number of infectious particles in viral stocks produced in HRT-18 cells (A), differentiated LA-N-5 cells (C), and murine primary CNS mixed cultures (E). For the evaluation of time of entry, HRT-18 cells (B) and differentiated LA-N-5 cells (D) were seeded in 24-well plates and incubated for 1 h on ice with either rOC/ATCC, rOC/SG758R, or rOC/SR903A at an MOI of 1. Then cells were incubated for 20, 30, and 50 min at 37°C, medium was replaced with new medium containing chloroquine (200 nM), and cells were incubated for 16 h at 37°C. Positive controls were incubated for 16 h without chloroquine. Viral entry was evaluated by immunofluorescence assay and quantified with CellProfiler software. Percentage of infection was set at 100% in control for each virus, and data at different time points were compared to this value. Tables below panels B and D represent the slope calculated over a 50-min time period. Differences were significant (*, P ≤ 0.05, **, P ≤ 0.01, and ***, P ≤ 0.001) or not significant (ns), and results shown are the mean values (with standard deviations) from three independent experiments.
The S glycoprotein may be cleaved by cathepsins B and L and by TMPRSS5 (transmembrane serine protease 5)/spinesin.
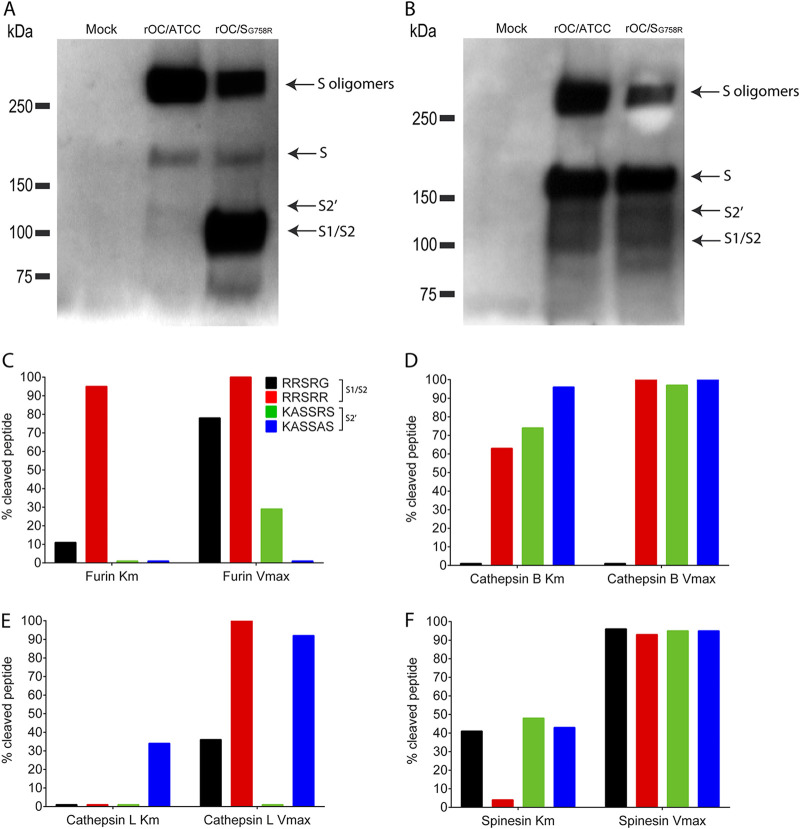
We next sought to evaluate the importance of the cleavage steps in infection. We first looked at the S protein processing in the LA-N-5 neuronal cell line and first confirmed previous results (55) showing that the cleavage of the S protein in S1/S2 fragments was mostly efficient for rOC/SG758R mutant virus at 48 h postinfection (hpi) (Fig. 3A) compared to the protein associated with cells (Fig. 3B). Our results also showed a very faint intermediate size band between the uncleaved and S1/S2 cleaved forms of the S protein for reference virus and rOC/SG758R mutant virus which we also already previously reported (55) (Fig. 3A and B, black arrow S2′). No data are presented for the rOC/SR903A virus because infection of human neuronal LA-N-5 cells is highly inefficient and consequently does not produce enough viral proteins (including S) to be detectable even at 48 hpi.
FIG 3.
Cathepsins B and L and TMPRSS5/spinesin are potential players in S glycoprotein cleavage. Differentiated LA-N-5 cells were infected with rOC/ATCC or rOC/SG758R at an MOI of 0.2. Proteins in association with cells or in supernatant were extracted at 48 hpi, and Western blot analysis of cell culture supernatant (A) or whole-cell lysates (B) (10 μg of proteins loaded in all wells as measured by BCA) revealed the presence of oligomers of S proteins (250 kDa), the uncleaved monomer form of the S glycoprotein (180 kDa), a cleaved form at about 100 kDa (S1/S2), and presumably the second cleaved site at about 120 kDa (S2′). Synthetic peptides represent all virus S protein at the S1/S2 site, RRSRG (rOC/ATCC; black) and RRSRR (rOC/SG758R; red), or at S2′ site, KASSRS (rOC/ATCC; green) and KASSAS (rOC/SR903A; blue). The different peptides were incubated with furin (C), cathepsin B (D) or L (E), or TMPRSS5/spinesin (F) for 30 min, and percent cleaved peptide was measured as described in Materials and Methods. Cleavage at Km is a representation of an early step of viral infection (small amount of peptide) and Vmax a late step of viral infection (large amount of peptide).
Cleavage at S2′ appears to occur mostly during viral entry and is likely transient in nature, rendering the phenomenon difficult to observe (51). Therefore, we sought to identify which proteases could potentially play a role in cleavage of the HCoV-OC43 S glycoprotein by designing synthetic peptides, described in Materials and Methods and representing all viruses at the S1/S2 site (RRSRG [rOC/ATCC] and RRSRR [rOC/SG758R]) (55) or at the S2′ site (KASSRS [rOC/ATCC] and KASSAS [rOC/SR903A]). The Km values of the graphs represent the situation when the enzymes are in the presence of a small amount of substrate (corresponding to beginning of infection), and the Vmax values represent the situation when the amount of substrate is high (corresponding to the end of the replicative cycle of the virus) (Fig. 3C to F). As shown in Fig. 3C, we confirmed previous results (55) showing that furin was effective at cleaving the peptides representing the S1/S2 site and that it was much more efficient for the peptide containing the rOC/SG758R virus sequence (RRSRR). Our new data clearly indicate that this proprotein convertase has a very low affinity for the putative cleavage site at S2′, especially for the Km portion (Fig. 3C). Cathepsin B was highly active at cleaving peptide containing the rOC/SG758R sequence (RRSRR) at both Km and Vmax but was unable to cleave peptide RRSRG (Fig. 3D), whereas cathepsin L was only able to partially cleave peptide KASSAS (mutant S2′ site) at Km while being much more effective at Vmax (Fig. 3E). We are aware that data obtained by such in vitro biochemical studies need to be interpreted with all prescribed caution. However, this method was previously used for studying the potential cleavage of SARS-CoV-2 S protein (79, 80), and our data confirmed previous results related to cleavage by furin-like proteases and presage that at the beginning of infection (entry), cathepsin B would be able to efficiently cleave the S glycoprotein at the S1/S2 and S2′ sites and that the cleavage by cathepsin L would be less efficient for all viruses. The Vmax values presage that cathepsins B and L would also be active and efficient later during infection but not equally for all viruses. Our data on TMPRSS5/spinesin suggest that virus rOC/SG758R would not use this protease for its cleavage at S1/S2 in the early steps of infection but that all viruses could use it at later stages (Fig. 3F).
S protein potential cleavage sites differentially influence entry in neuronal cells.
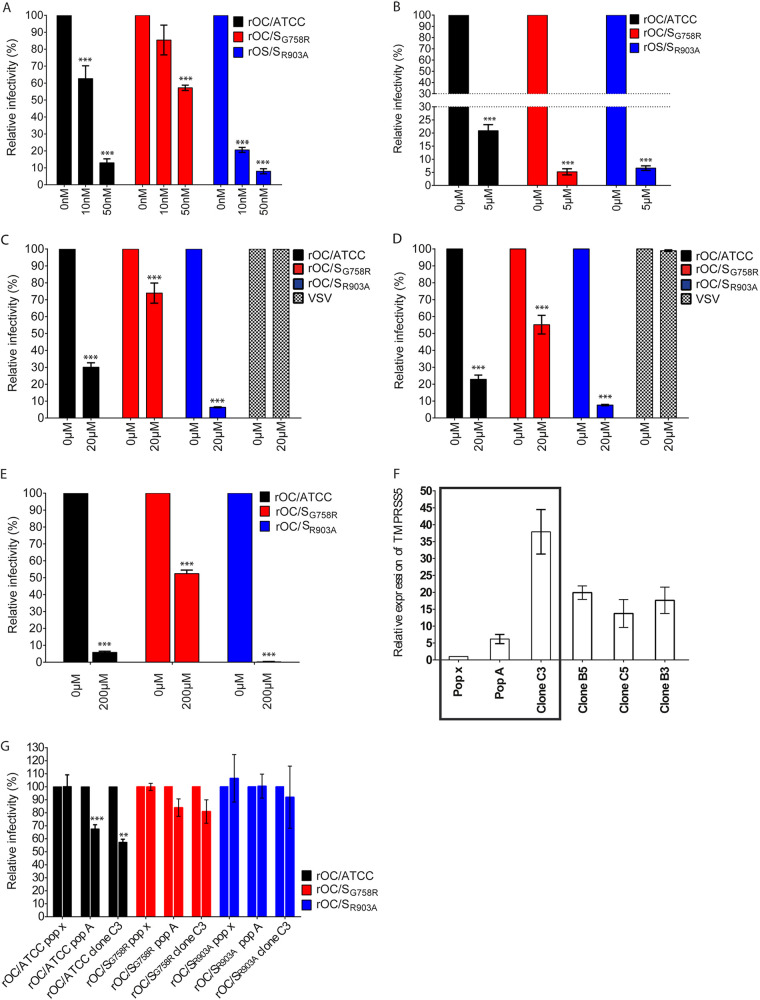
Having shown that cathepsins and TMPRSS5/spinesin are potential host proteases able or not to cleave peptides representing the S protein S1/S2 and S2′ sites (Fig. 3) and knowing that these types of proteases (members of cathepsins and TMPRSSs) can be involved in early steps of infection (entry) for many viruses, including CoVs (51, 56, 57, 60, 61, 74, 75, 81, 82), we evaluated if they could be involved during infection of neuronal cells by our different viruses. As cathepsins are predominantly and typically found in endosomes (cathepsin B) and lysosomes (cathepsin L), where they are optimally active at low pH, and as the endocytic path was shown to be used by CoVs (83–85), we evaluated the importance of these proteases during endocytosis. We first evaluated viral entry, with chloroquine. Entry of mutant rOC/SR903A virus was mostly affected in LA-N-5 cells in a dose-dependent manner when endosomal acidification was neutralized. Indeed, 10 nM chloroquine significantly decreased mutant rOC/SR903A entry, by 80%, whereas this reduction was 40% for rOC/ATCC and not significant for rOC/SG758R. These results indicate a difference in sensitivity to chloroquine and suggest that the mutant G758R is less dependent on acidification and low-pH-active proteases. As S protein mutants arise during infection by coronaviruses, including SARS-CoV-2, this suggests that variants such as the G758R mutant may represent chloroquine-resistant mutants that may arise during the course of infection. With a higher concentration of chloroquine (50 nM), all three variants were affected (Fig. 4A). Moreover, acidification of endosomes was also demonstrated to be necessary for viral entry in primary CNS cultures of mice (Fig. 4B). These data suggest that the rOC/SG758R virus, harboring mostly an already cleaved S protein at the S1/S2 site (55) (Fig. 3), relies less on endosome/lysosome acidification to enter cells.
FIG 4.
Endosomal acidification and proteases are necessary for efficient entry of HCoV-OC43 into neuronal cells. Unless stated otherwise, infections were performed with either rOC/ATCC, rOC/SG758R, or rOC/SR903A at an MOI of 1 and cells were incubated for 16 h at 37°C and each virus is compared to itself under all different conditions. Thus, for each virus, infection without inhibitor is set at 100%. (A and B) Differentiated LA-N-5 cells (A) or mixed primary cultures of mouse brain (B) were incubated as described in Materials and Methods with the indicated concentration of chloroquine. (C and D) Differentiated LA-N-5 cells were incubated with 20 μM MDL 28170 (C) or 20 μM Z-FA-FMK (D). Infections were performed with either rOC/ATCC, rOC/SG758R, rOC/SR903A, or VSV at an MOI of 1. After incubation with inhibitors for 2 h, medium was removed, fresh medium with 200 nM chloroquine was added, and cells were put back at 37°C for another 16 h. (E) Mixed primary cultures of mouse brain incubated with 200 μM camostat mesylate were infected with either rOC/ATCC, rOC/SG758R, or rOC/SR903A at an MOI of 0.2 for 16 h at 37°C. After fixation, viral antigens were stained with rabbit antiserum against the S viral glycoprotein and neurons with a monoclonal antibody (MAb) against MAP2. Percentage of infection was set at 100% in control (0 μM) for each virus. (F) Evaluation of TMPRSS5/spinesin levels of expression in different puromycin-selected populations and isolated clones of LA-N-5 cells transduced by lentiviral vector. Boxed populations and clones were evaluated for panel G. (G) Differentiated LA-N-5 cells ectopically expressing TMPRSS5 (pop A and clone C3) or LA-N-5 control (pop X) were incubated with 200 μM camostat mesylate and were infected with virus rOC/ATCC, rOC/SG758R, or rOC/SR903A at an MOI of 0.2 for 16 h at 37°C. Viral entry was evaluated by immunofluorescence assay and quantified with CellProfiler software. Results are the mean values (with standard deviations) from three independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Having shown that endosomal acidification influenced entry (Fig. 4) and that cathepsins could potentially be involved in the process (Fig. 3), we sought to evaluate the implication of these endosomal proteases during infection. For this, we infected LA-N-5 cells in the presence of either MDL-28170 (chemical inhibitor of cathepsin B [Fig. 4C]) or Z-FA-FMK (chemical inhibitor of both cathepsin B and L [Fig. 4D]). In both cases, entry of rOC/ATCC and rOC/SR903A mutant virus (which both harbor a suboptimal cleavage site RRSRG at S1/S2) was mostly altered compared to that of rOC/SG758R virus (Fig. 4C and D). All these data suggest that the 3 viruses use the endosomal pathway to enter neuronal cells and that cathepsins have a differential impact depending on the presence or absence of optimal S1/S2 or S2′ sites. Vesicular stomatitis virus (VSV) was used as a negative control, as its glycoprotein G does not need to be cleaved by cellular proteases within the endocytic pathway to enable virus entry (82). Endocytosis (pH and cathepsin inhibition) experiments on epithelial HRT-18 cells also revealed that the three viruses partly rely on this pathway for infection of epithelial cells (data not shown). However, inhibitors of cathepsins were highly cytotoxic on CNS primary cultures even at low doses and thus could not be tested for inhibition of infection.
Other host proteases, such as members of the membrane-bound TMPRSSs, may also influence coronavirus infection (70, 82, 86), supposedly by initiating direct membrane fusion at the cell surface and subsequent viral entry in susceptible cells (51, 74, 76, 82, 87). To first investigate if TMPRSSs are involved in HCoV-OC43 entry in neuronal cells, we used the chemical inhibitor camostat mesylate, and no difference in efficiency of entry between viruses was observed in LA-N-5 cells. RT-PCR revealed that TMPRSS2 and -5 were expressed below the limit of detection in LA-N-5 cells but that TMPRSS5/spinesin, a form of TMPRSS usually expressed in CNS cells, was highly expressed in murine primary CNS cultures (data not shown). We thus tested the implication of TMPRSS5/spinesin in these primary cultures, by performing infection in the presence of camostat mesylate (51) and found that entry of rOC/ATCC and rOC/SR903A viruses was more affected than rOC/SG758R virus (Fig. 4E). These data strongly suggest that viruses that harbor a suboptimal cleavage S1/S2 site (RRSRG; rOC/ATCC and rOC/SR903A) rely more on cleavage by TMPRSS (most probably TMPRSS5 in the CNS) for efficient entry, and again, the modification of the wild-type S2′ site KASSRS (rOC/ATCC) to KASSAS (as in the rOC/SR903A virus) does not prevent this potential cleavage and eventual associated entry path, such as the proposed fusion at the cell membrane. Considering that camostat mesylate had a significant effect on viral infection in primary cells (Fig. 4E), which express TMPRSS5/spinesin, we decided to ectopically express this host protease in LA-N-5 cells and monitor its effect at early stages of viral infection. Ectopic expression in the presence of camostat mesylate significantly reduced infection by reference rOC/ATCC in a dose-dependent manner and had a lower effect during infection by the other two viruses (Fig. 4F and G).
The arginine in the putative S2′ site in the S glycoprotein is necessary to produce infectious viral particles that efficiently propagate in neuronal cell culture.
To push further our investigation on the role of the S2′ site during HCoV-OC43 infection, we evaluated the kinetics of viral replication and spreading in cell culture. Even though a slight difference in replication and spread was observed between all three viruses in HRT-18 human epithelial cells (data not shown), statistical analysis revealed that it was not significant. On the other hand, even though both rOC/ATCC and rOC/SG758R variants were able to spread and produce infectious particles in LA-N-5 human cells and in murine mixed primary CNS cultures (Fig. 5), the mutant rOC/SR903A was heavily restricted for both parameters of infection (Fig. 5A to D). However, ectopic expression of TMPRSS5/spinesin increased production of infectious virus and propagation for all three viruses in LA-N-5 cells (Fig. 5E and F). Moreover, we previously showed that HCoV-OC43 spread in neuronal cell cultures could partially be attributed to axonal transport associated with a cell-to-cell mode of propagation (88). Using both LA-N-5 cells and murine primary CNS cultures in both fluid (cell-free propagation of viral particles in medium) and semifluid/methylcellulose (cell-to-cell propagation), we thus evaluated the relative spreading capacities of all three viruses and found that mutant rOC/SR903A cell-to-cell propagation was almost totally abrogated in both LA-N-5 cells and murine primary cells (Fig. 5G to J).
FIG 5.
The wild-type S2′ site in S glycoprotein is necessary to efficiently produce infectious viral particles and for efficient propagation in neuronal cell cultures. Differentiated LA-N-5 cells or mixed primary cultures were infected with rOC/ATCC, rOC/SG758R, or rOC/SR903A at an MOI of 0.2 or 0.03, respectively. Kinetics of infectious virus production (A) and viral spreading (B) in LA-N-5 cells, mixed primary cultures from mouse brain (C and D), and LA-N-5 cells ectopically expressing TMPRSS5 (E and F) was determined. LA-N-5 cells (G and H) or mixed primary cultures from mice brain (I and J) were infected (MOI of 0.01) and then overlaid with fluid or semifluid medium for 16 to 72 h as described in Materials and Methods. Cells were then fixed and immunostained. For LA-N-5 cells, MAb against the S glycoprotein and DAPI for cell nucleus were used, and for mixed primary cultures, rabbit antiserum against the S glycoprotein, MAb against the neuronal protein MAP2, and DAPI for cell nucleus were used. The propagation efficiency was plotted as the ratios between the percentages of infected cells at 72 h versus 16 h. Differences were significant (*, P ≤ 0.05, **, P ≤ 0.01, and ***, P ≤ 0.001), and results are the mean values (with standard deviations) from three independent experiments. Cells were counted with CellProfiler software. LoD, limit of detection.
Mutation at both S1/S2 and S2′ sites reduces infection and early propagation among olfactory sensory neurons (OSN).
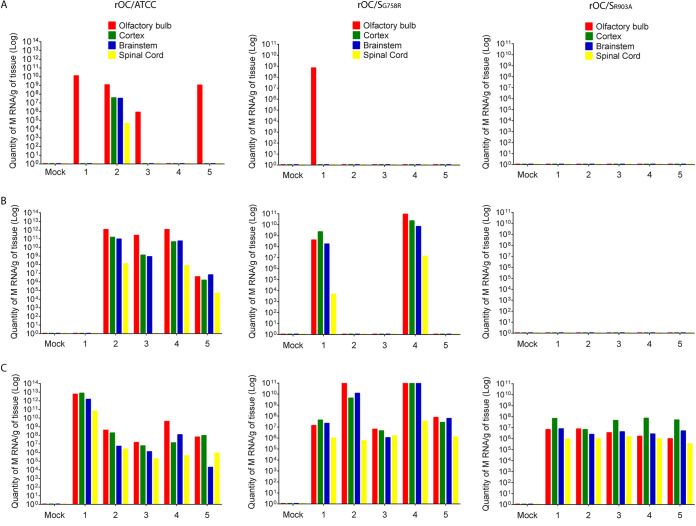
We previously reported that cleavage of the HCoV-OC43 S protein at the S1/S2 site by host cell proteases was not essential for neuroinvasion but was an important determinant of virus spread in the CNS. Therefore, 10-day-old C57BL/6 mice were inoculated intranasally (i.n.) and the presence of viral RNA was evaluated in olfactory bulb, cortex, brain stem, and spinal cord to compare the levels of neuroinvasion and propagation within the CNS between the reference virus and both single mutant viruses (Fig. 6). Viral RNA was first detected in the olfactory bulb and later in other structures of the mouse CNS, suggesting that neuroinvasion per se was not altered for any virus. However, the reduction in the number of infected mice, the delay in RNA detection, and the lower RNA copy number of rOC/SR903A may underline a defect in early propagation in the CNS via by the olfactory route.
FIG 6.
Mutation at either S1/S2 or S2′ site delays viral neuroinvasiveness and propagation within discrete regions of the brain. Ten-day-old C57BL/6 mice received 103.5 TCID50/10 μl of rOC/ATCC, rOC/SG758R, rOC/SR903A, or PBS by the i.n. route. Different regions of the brain were harvested at 2 (A), 3 (B), and 4 (C) dpi, and viral RNA was quantified by qRT-PCR. Each bar (x axis numbers 1 to 5) represents one single infected mouse, and “mock” represents a mock-infected mouse.
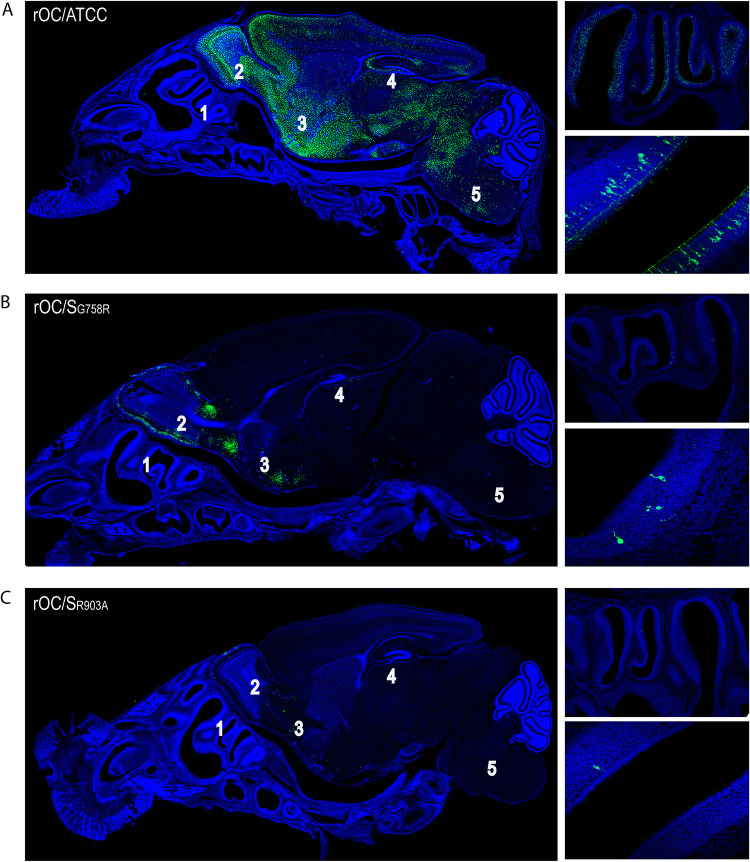
To determine whether these variations in viral RNA content could result in a difference in cell type infection in the olfactory epithelium, we performed an immunofluorescence assay at 3 days postinfection (dpi) on mouse decalcified whole heads after i.n. inoculation. Our data correlated with the amount of viral RNA found in the mouse CNS for all 3 viruses. Indeed, rOC/ATCC (Fig. 7A, left image) was able to efficiently invade and spread in the CNS, and conversely, both mutants (Fig. 7B and C, left images) presented a defect in their infection capacities. Reference virus rOC/ATCC was clearly the most efficient virus to infect and spread among OSN in the olfactory epithelium (Fig. 7A, right images, which are higher magnifications of the left image), followed by rOC/SG758R and rOC/SR903A mutants, respectively (Fig. 7B and C, right images, top and bottom). Whereas these data could presage a relative defect in neuroinvasion for mutants particularly important for rOC/SR903A, the observation that fewer OSN were infected may rather reveal a decreased capacity to propagate between the OSN in the nasal cavity, similar to the delay observed in neuronal cell cultures (Fig. 5). Together, these data clearly indicate that S1/S2 and mostly S2′ sites in the S protein play a crucial role during HCoV-OC43 infection of the CNS.
FIG 7.
Mutation at either the S1/S2 or S2′ site reduces infection of olfactory sensory neurons (OSN) and delays viral neuroinvasiveness and propagation within discrete regions of the brain. Ten-day-old C57BL/6 mice received 103.5 TCID50/10 μl of rOC/ATCC, rOC/SG758R, or rOC/SR903A by the i.n. route, and histological examination of decalcified whole head was performed to visualize virus spread in the CNS at 3 dpi for rOC/ATCC (A), rOC/SG758R (B), or rOC/SR903A (C). Detection of viral antigens (green; MAb against the S viral glycoprotein) and cell nucleus (DAPI) is presented on the left. Pictures at the top right represent olfactory epithelium, and pictures at the bottom right represent higher magnifications of infected olfactory sensory neurons. Magnifications are ×10 for left images, ×20 for upper right images, and ×63 for lower right images. 1, nasal cavity; 2, olfactory bulb; 3, pyriform cortex; 4, hippocampus; 5, brain stem.
The rOC/SR903A mutant virus is avirulent compared to rOC/ATCC and rOC/SG758R.
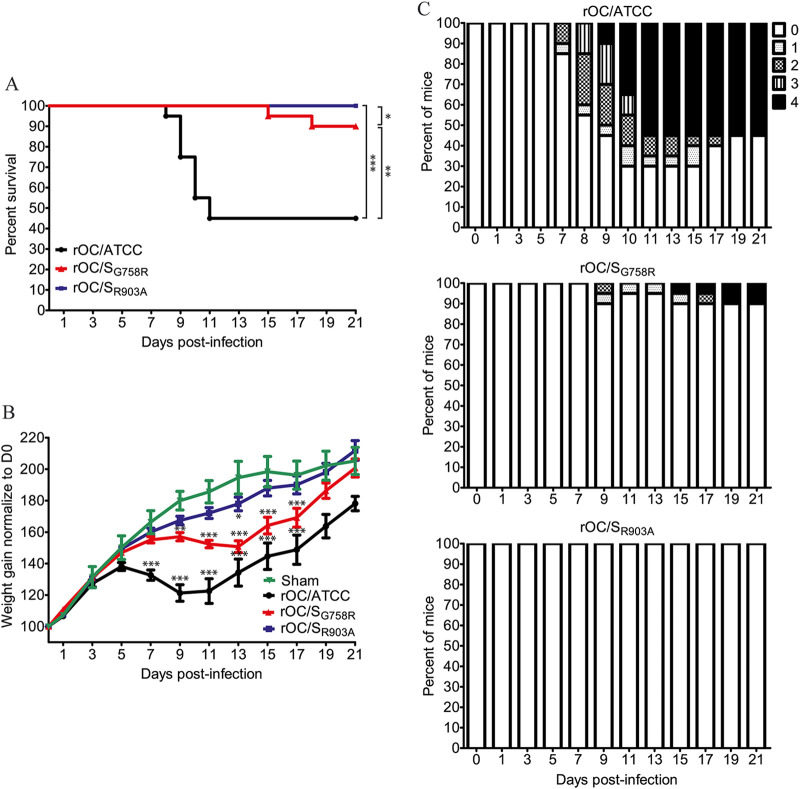
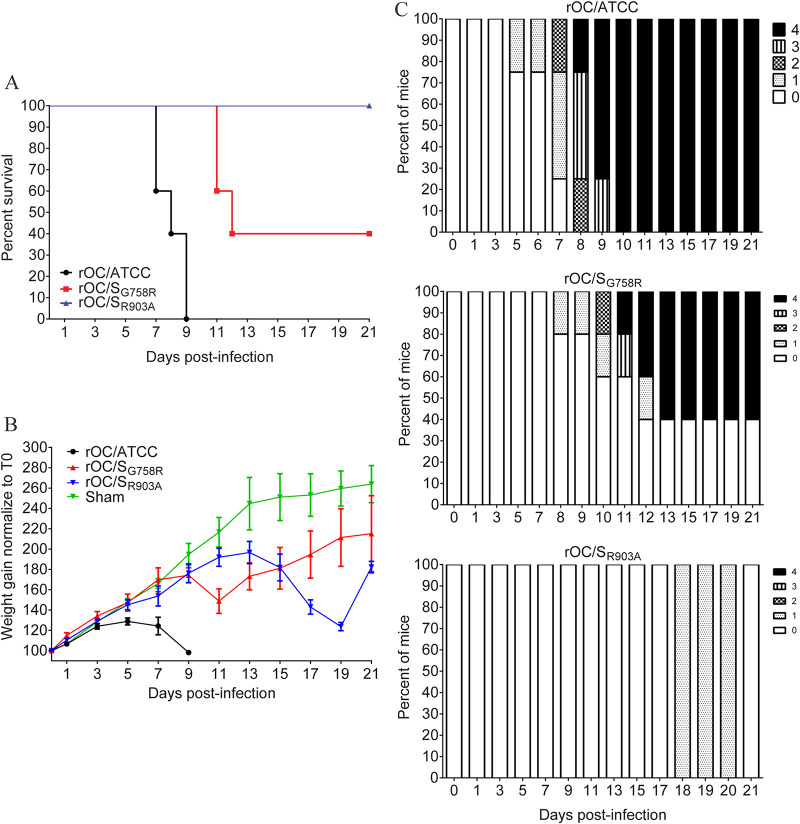
Intranasal injections of virus result in infection of the CNS only 68% of the time; therefore, we decided to evaluate the different parameters of infection using intracranial (i.c.) injection, a method by which 100% of the mice are efficiently infected with the same propagation pathway within the brain (88). Therefore, C57BL/6 mice were infected by i.c. injection and their survival, neurological symptoms, and weight gain were assessed as previously described (55). There was a significant difference in survival after inoculation of either virus (Fig. 8A): like the sham control, the rOC/SR903A virus induced no mortality compared to rOC/SG758R and rOC/ATCC, which induced 10% and 55% mortality, respectively. On the other hand, there was a significant delay in body weight gain with rOC/ATCC and the mutant rOC/SG758R virus compared to rOC/SR903A and the sham control (Fig. 8B). Using the clinical score scale previously described (55), we next studied the neurological symptoms of mice. Mice infected by the mutant rOC/SR903A virus, or by the sham control, did not develop any clinical sign. Conversely, mice infected by the mutant rOC/SG758R developed the 4 levels of symptoms, but less frequently (10%) than with reference rOC/ATCC, which induced encephalitis associated with the 4 different levels of clinical score more frequently (70% of infected mice) (Fig. 8C). Taken together, survival and weight curves coupled with the clinical scores indicated that the mutant virus rOC/SR903A was totally avirulent.
FIG 8.
The rOC/SR903A mutant virus is avirulent in C57BL/6 mice. Twenty-two-day-old C57BL/6 mice received 101.5 TCID50/10 μl of rOC/ATCC, rOC/SG758R, rOC/SR903A, or PBS by the i.c. route. (A) Survival curves of mice over a 21-day period. Differences between the three viruses were significant compared to sham control and to each other (*, P ≤ 0.05, **, P ≤ 0.01, and ***, P ≤ 0.001). (B) C57BL/6 mice were weighed every 2 days over a 21-day period to estimate weight variations, which were expressed as percentages, compared to day 0 (100%). Differences were significant between 7 and 17 dpi, when the three conditions (rOC/ATCC, rOC/SG758R, and rOC/SGR903A) were compared to sham control. (C) Evaluation of the clinical scores (percentage of mice at each level of the scale) of mice infected by rOC/ATCC, rOC/SG758R, or rOC/SGR903A based on neurological symptoms described in a clinical score scale between level 0 and 4 (see Materials and Methods).
Viral propagation and replication in the CNS of infected mice are reduced for mutant rOC/SR903A virus compared to rOC/SG758R and reference wild-type virus.
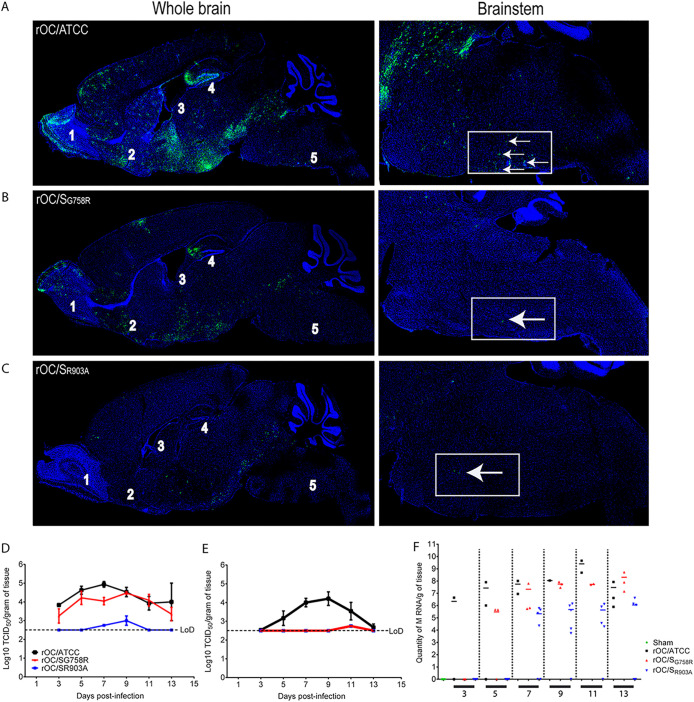
In order to decipher the mechanisms associated with the differential neurovirulence, i.c. injections were again performed with all three HCoV-OC43 variants. Even though the infected regions were similar for all 3 viruses, spread of the mutant rOC/SR903A virus was abrogated compared to that of rOC/ATCC and rOC/SG758R. Indeed, whereas rOC/ATCC can already spread in almost all regions of the brain at 7 dpi (Fig. 9A, left image) and is able to travel through the brain stem to reach the spinal cord (Fig. 9A, right image), mutant viruses rOC/SG758R and rOC/SR903A were both less efficient in spreading within different regions of the brain, as fewer viral antigens were detected, especially in the brain stem, where only scarce cells appeared infected (Fig. 9B and C, left images).
FIG 9.
rOC/SR903A mutant virus spread in the CNS is abrogated compared to that of rOC/ATCC and rOC/SG758R. (A to C) Histological examination of virus spread within the brains of 22-day-old C57BL/6 mice infected with 101.5 TCID50/10 μl of rOC/ATCC (A), rOC/SG758R (B), or rOC/SR903A (C) by the i.c. route. Detection of viral antigens in whole brain (left) or in the brain stem (right) of infected mice at 7 dpi is shown. White arrows represent infected cells that were stained in green with MAb against the S viral glycoprotein. Blue staining is DAPI staining the nucleus (DNA). Magnifications are ×10 for left images and ×20 for right images. 1, olfactory bulb; 2, pyriform cortex; 3, lateral ventricle; 4, hippocampus; 5, brain stem. (D and E) Production of infectious viral particles was measured in brains (D) and spinal cords (E) every 2 days between 3 and 13 dpi. Results are the mean values (with standard deviations) from three independent experiments. (F) In spinal cord samples where no infectious virus was detected, RNA of each virus was quantified by qRT-PCR between 3 and 13 dpi after i.c. injection in C57BL/6 mice. Every symbol represents one single infected mouse.
Given our observations on in vivo neuropropagation (Fig. 9), we evaluated whether this correlated with a difference in viral replication in the CNS. Brains and spinal cords were harvested and infectious virus production was assayed every 2 days for a period of 21 days. A very important reduction in the amount of infectious viral particles in the brain was observed for rOC/SR903A, and there was a drastic difference in the production of infectious virus in the spinal cord for both mutants compared to the rOC/ATCC. Infectious titers of rOC/ATCC were almost identical to those detected in the brain, whereas the less virulent mutant rOC/SG758R (55) and the avirulent rOC/SR903A apparently poorly reached the spinal cord, as infectious virus particles were below the limit of detection in almost all of the 30 infected mice (Fig. 9D and E). In order to determine if the mutant viruses were able to reach spinal cord even in the absence of detectable infectious virus in the vast majority of mice, we investigated for the presence of viral RNA in all spinal cord samples where no infectious viral particles had been detected between 3 and 13 dpi. Viral RNA was present for all three viruses, although there was a delay for the mutant viruses. Indeed, rOC/ATCC RNA was detected as early as 3 dpi, whereas viral RNA was first detected only at 5 and 7 dpi for mutants rOC/SG758R and rOC/SR903A, respectively (Fig. 9F), indicating an important defect in viral propagation within mice CNS that correlates with the very rare detection of infectious virus.
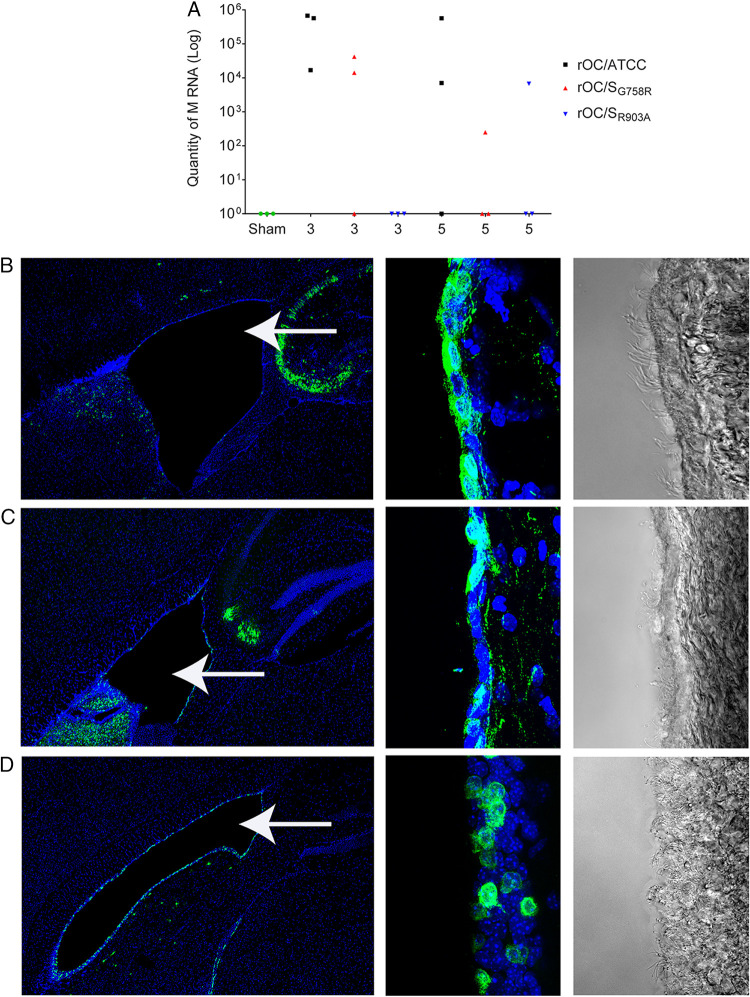
HCoV-OC43 capacity to enter CSF and reach the spinal cord is not related to S protein S1/S2 and S2′ sites and is not influenced by infection of ependymal cells.
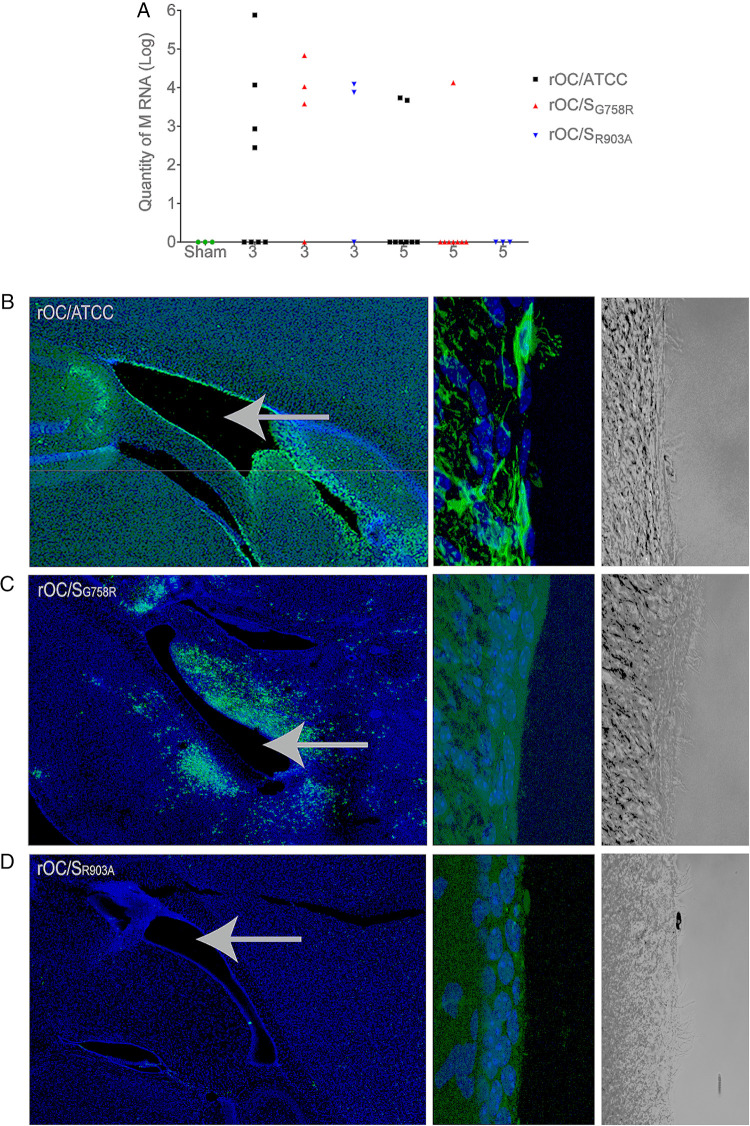
As both mutant viruses were able to reach the spinal cord (viral RNA detected [Fig. 9F]), despite an apparent inefficiency to achieve that through the brain stem and to produce detectable infectious virus, we wished to evaluate if cerebrospinal fluid (CSF) could be used for further spreading toward spinal cord as described for other neurotropic viruses (89, 90). Between 3 and 5 dpi, all 3 viruses were found at equivalent levels in the CSF (Fig. 10A). We also harvested brains at 5 dpi for histological observation and noticed that rOC/ATCC virus could very efficiently infect nonneuronal cells surrounding the lateral ventricles (Fig. 10B, left image) conversely to both mutant rOC/SG758R and rOC/SR903A viruses, which barely infected these cells (Fig. 10C and D, left image). A higher magnification allowed us to identify these cells as rOC/ATCC virus-infected ependymal cells (Fig. 10B, right image, and data not shown) and confirmed that both mutants were scarcely detected in these nonneuronal cells (Fig. 10C and D). These results presage another route of propagation for reference virus (rOC/ATCC) within the CNS, which may suppose a change in cell tropism, as both mutant viruses almost completely lost their ability to infect ependymal cells.
FIG 10.
Viral RNA of all variants is present in CSF, but ependymal cells are efficiently infected only by reference virus rOC/ATCC. Twenty-two-day-old C57BL/6 mice received 101.5 TCID50/10 μl of rOC/ATCC, rOC/SG758R, rOC/SR903A, or PBS by the i.c. route. (A) RNA of each virus was quantified by qRT-PCR at 3 and 5 dpi in CSF of C57BL/6 mice after i.c. injection. Every symbol represents one single infected mouse. (B to D) Histological examination of virus spread after 5 dpi. (B) rOC/ATCC; (C) rOC/SG758R; (D) rOC/SR903A. Sections of brains were stained with MAb against the S viral glycoprotein (green). Blue is DAPI, which stains the nucleus (DNA). Left images represent lateral ventricles (white arrows) and surrounding region in mouse brain (magnification, ×10), and right images represent higher magnification (63×) of ependymal cells surrounding ventricles seen in immunofluorescence and bright field, respectively.
Innate immunity reduces propagation in the CNS and associated neurovirulence for all three HCoV-OC43 variants.
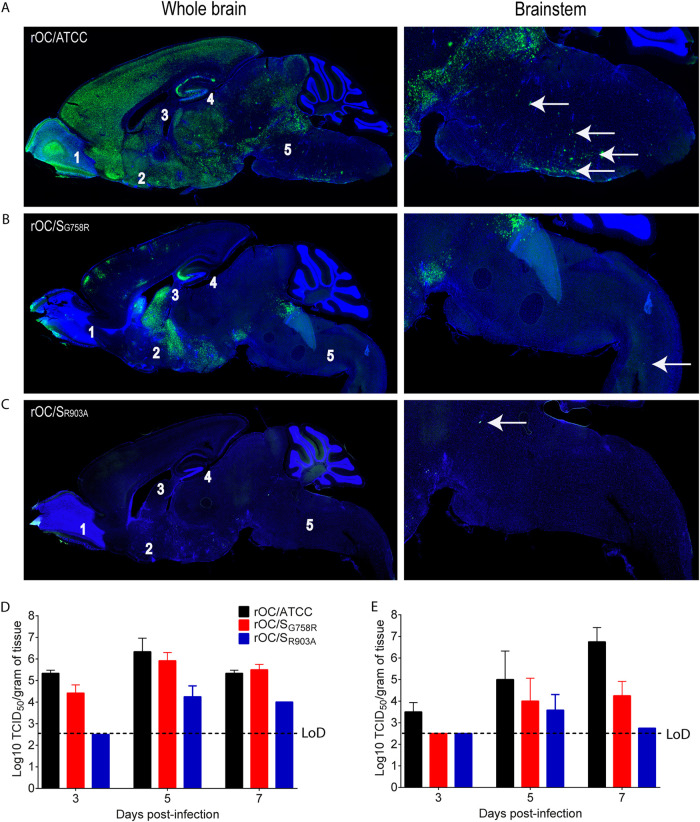
We previously showed that innate immunity (astrogliosis and microgliosis) was induced differentially by diverse HCoV-OC43 variants in direct correlation with viral propagation and neurovirulence (38, 55, 91). Therefore, we next sought to compare the relative importance of innate immunity in controlling CNS infection by our three HCoV-OC43 variants (harboring a potentially differentially cleaved S protein). Making use of IFNAR knockout (IFNAR-KO) mice, we examined if the differences between the three variants would remain, regardless of innate immunity status. Intracranial injection of all three variants into IFNAR-KO mice revealed an increased neurovirulence for rOC/ATCC and rOC/SG758R, but with the exception of a weight gain defect (compare Fig. 11B to Fig. 8B), the phenotype (clinical scores and survival) of rOC/SR903A remained the same as what was observed in immunocompetent C57BL/6 mice. Even though the differences in viral propagation between all three variants also remained similar to what was observed in C57BL/6 mice (compare Fig. 12A to C to Fig. 9), higher levels of infectious viruses were detected in the spinal cord of IFNAR-KO mice than of C57BL/6 mice infected by rOC/SG758R and rOC/SR903A (compare Fig. 12D and E to Fig. 9D and E). Interestingly, this increase in infectious virus production was associated with a modification of cell tropism, as in immunocompromised mice, both mutants were able to infect ependymal cells almost as efficiently as reference rOC/ATCC (compare Fig. 13 to Fig. 10).
FIG 11.
The rOC/SR903A mutant virus remains avirulent even in the absence of an adequate IFN-1 response. Twenty-two-day-old IFNAR-KO mice received 101.5 TCID50/10 μl of rOC/ATCC, rOC/SG758R, rOC/SR903A, or PBS by the i.c. route. (A) Survival curves of mice over a 21-day period. (B) IFNAR-KO mice were weighed every 2 days over a 21-day period to estimate weight variations, which were expressed as percentages, compared to day 0 (100%). (C) Evaluation of the clinical scores (percentage of mice at each level of the scale) of mice infected by rOC/ATCC, rOC/SG758R, or rOC/SGR903A based on neurological symptoms described in a clinical score scale between level 0 and 4 (see Materials and Methods).
FIG 12.
Differences between kinetics of viral propagation remain the same in the brains of IFNAR-KO mice, but both mutant viruses produce detectable levels of infectious virus in the brain and spinal cord. (A to C) Histological examination of virus spread within the CNS of 22-day-old IFNAR-KO mice infected with 101.5 TCID50/10 μl of rOC/ATCC (A), rOC/SG758R (B), or rOC/SR903A (C) by the i.c. route. Detection of viral antigens in whole brain (left images) or in the brainstem (right images) of infected mice at 7 dpi is shown. White arrows represent infected cells that were stained in green with MAb against the S viral glycoprotein. Blue is DAPI, which stains the nucleus (DNA). Magnifications are ×10 for left images and ×20 for right images. 1, is olfactory bulb; 2, pyriform cortex; 3, lateral ventricle; 4, hippocampus; 5, brain stem. (D and E) Production of infectious viral particles was measured in brains (D) and spinal cords (E) every 2 days between 3 and 7 dpi. Results are the mean values (with standard deviations) from three independent experiments.
FIG 13.
All three variants are present in CSF and replicate in ependymal cells of IFNAR-KO mice. Twenty-two-day-old IFNAR-KO mice received 101.5 TCID50/10 μl of rOC/ATCC, rOC/SG758R, rOC/SR903A, or PBS by the i.c. route. (A) RNA of each virus was quantified by qRT-PCR between 3 and 7 dpi in CSF. Every symbol represents one single infected mouse. (B to D) Histological examination of virus spread after 5 dpi. (B) rOC/ATCC; (C) rOC/SG758R; (D) rOC/SR903A. Sections of brains were stained with MAb against the S viral glycoprotein (green). Blue is DAPI, which stains the nucleus (DNA). Left images represent lateral ventricles (white arrows) and surrounding region in mouse brain (magnification, ×10) and right images represent higher magnification (×63) of ependymal cells surrounding ventricles seen in immunofluorescence and bright field, respectively.
DISCUSSION
Like the newly discovered SARS-CoV-2, the human coronavirus OC43 (HCoV-OC43) is an opportunistic pathogen, with potential neuroinvasive and neurotropic properties, which raises interest in studying its potential involvement in neurological disease (26, 36, 39, 44, 55, 92–94).
Activation of viral envelope glycoproteins like coronaviruses S glycoprotein by host cell proteases is often a critical step during infection, and modulation of this process can have a profound effect by influencing early (entry) and late stages of viral infection and cell-cell transmission and potentially in modulation of cell tropism (51–54, 56, 57, 59, 60, 74–76, 85, 95–99). Having previously shown that HCoV-OC43 S protein cleavage is involved in CNS infection, we herein describe a novel feature of HCoV-OC43 S protein, which better characterizes the importance of the process from entry to propagation between CNS cells, and show that this process also involves adequate IFN-1 response.
During entry, coronaviruses can use acid-dependent late endosome-associated cathepsins (54, 61, 81, 84, 85, 96, 100–102), furin (early endosome) (62, 103), and TMPRSS2, at or near the cell surface (56, 57, 59, 74, 76, 85, 101, 104–106). Therefore, the S2′ putative cleavage site in the OC43 S protein could be used by these cell proteases (51, 60, 107, 108) to participate in the complete activation of the S protein. As previously mentioned, our data obtained by in vitro biochemical studies need to be interpreted with all prescribed caution, as peptide may not resemble the “real” conformation of the complete S protein. However, this method was previously used for studying the potential cleavage of SARS-CoV-2 S protein (79, 80), and our data confirmed previous results related to cleavage by furin-like proteases (55). Interestingly, cathepsin B was able to cleave all peptides except peptide 322 (RRSRR site), and cathepsin L cleaved more efficiently the KASSAS peptide (corresponding to mutated putative S2′ site) than the reference sequence peptide (KASSRS). This suggests that the latter protease would not play an important role in the endosomal pathway during infection by naturally circulating HCoV-OC43 variants detected in clinical isolates, which all possess the KASSRS sequence at the S2′ site. On the other hand, our data also presage that this member of the cathepsin family may rather play an important role at a later stage of infection (Fig. 3E, Vmax data) on the S1/S2 site (RRSRR), also present in all HCoV-OC43 known clinical isolates. These data corroborate what was observed for other coronaviruses, including the newly identified SARS-CoV-2 (57, 81, 84, 85), for which cathepsins allowed efficient cleavage and activation of S glycoprotein. Moreover, reference virus rOC/ATCC S2′ site is critical for fast and optimal entry of neuronal cells by HCoV-OC43 (Fig. 2). These results are reminiscent of data on MERS-CoV, for which a circulating variant harboring a mutation at the critical P1′ cleavage position in the S2′ site modulates spike-mediated fusion and viral entry into cells (109). Mutant rOC/SG758R was the least affected by cathepsin inhibitors (Fig. 4), possibly because its S protein is already cleaved at S1/S2. This was previously demonstrated for the murine coronavirus (mouse hepatitis virus [MHV]), for which a cathepsin-related cleavage was important for strain MHV-2 but not for strain A59 (81), the latter being cleaved at S1/S2.
TMPRSSs may also serve in entry for MERS-CoV, SARS-CoV, and SARS-CoV-2 (56, 59, 85, 99, 101), as well as for circulating HCoV (76, 86, 110). Camostat mesylate significantly decreases infection in murine CNS primary cells and in LA-N-5 cells, which ectopically express TMPRSS5 (Fig. 4), suggesting that HCoV-OC43 S protein may also be activated by members of the TMPRSS family, including the CNS-expressed TMPRSS5/spinesin. The mutant virus rOC/SG758R was the least affected by the TMPRSS inhibitor. Together with data presented in Fig. 3, this may underscore that cleavage by furin-like proteases at the optimal S1/S2 site in the S protein of rOC/SG758R (Fig. 3) (55) modifies the three-dimensional (3D) structure of the glycoprotein, rendering the S2′ site poorly accessible to TMPRSSs, or alters the virus-cell interaction in a way that these proteases become less efficient in cleaving the rOC/SG758R virus S protein during viral entry into the cells. Moreover, the observations that wild-type rOC/ATCC appears less sensitive to chloroquine inhibition in CNS primary cultures (Fig. 4B) and is heavily inhibited by camostat mesylate in these cells (Fig. 4E) and in LA-N-5 cells, which ectopically express spinesin (Fig. 4G), also suggest that it can use TMPRSS5 to activate its fusion to the cell membrane to infect the cells. Studies are under way to validate these potential cleavage-related results with full-length proteins and during infection with pseudotyped particles (111, 112).
In agreement with our previous results (55), the spreading of the rOC/SG758R mutant virus was delayed, but our new data underline that the S2′ site of the S glycoprotein is much more important for efficient spreading in cell culture and in mice. These results are reminiscent of data on porcine epidemic diarrhea coronavirus (PEDV) and SARS-CoV, for which introduction of a cleavage site in S2′ sequence positively impact viral replication in cell culture (61, 113). In addition to their possible role in viral entry, TMPRSSs have been shown to be involved in later stages of infection and virus release for other coronaviruses (51, 110, 114). Altogether, even though we did not show that cleavage of the S protein at the S2′ site was essential during infection per se, our observations strongly suggest that the S2′ spike cleavage (by TMPRSSs and/or cathepsins) is a prerequisite for efficient cell-cell propagation. Moreover, the suboptimal S2′ cleavage site (KASSAS) renders HCoV-OC43 totally avirulent (absence of neurological symptoms and mortality) and the corresponding rOC/SR903A variant is associated with an important defect in replication in both the brain and spinal cord (Fig. 6, 9, and 10).
Different pathways of neuroinvasion have been suggested for human coronaviruses, including the olfactory pathway, which could be related to the symptoms of dysosmia reported for a significant portion of SARS-CoV-2-infected individuals (14, 88, 94, 115–117). Even though the main cell target appears to be different than SARS-CoV-2 (93, 94), like this novel coronavirus, all three HCoV-OC43 variants used in our study invade the CNS through the olfactory epithelium (Fig. 7). The decreased efficiency of OSN infection by S1/S2 and S2′ HCoV-OC43 mutants may indicate a problem with viral entry and/or with neuropropagation between susceptible cells for mutant viruses rather than a change in cellular tropism in the first step of neuroinvasion in contrast to what has been previously reported for MERS-CoV infecting other cell types (75).
Although spread toward the spinal cord was delayed for both mutant viruses, they were able to reach it, as revealed by the presence of viral RNA after infection (Fig. 6 and 10). As all 3 viruses were also detected in CSF, the difference in spreading through neuronal cells in the brain stem may partially explain the differential capacity to reach the spinal cord and produce high levels of infectious virus associated with neurovirulence. The presence of viruses in the CSF is a typical feature of a viral CNS infection and has been suggested as a potential route of dissemination toward the spinal cord for other viruses, including SARS-CoV-2 (44, 89, 90, 94, 118–120). The presence of all 3 HCoV-OC43 variant RNAs in the CSF may indicate a plausible pathway of spread from the brain to the spinal cord. However, as there was no significant difference in the relative amounts of RNAs, this pathway probably does not account for the differential capacities of propagation toward spinal cord between the three viruses. On the other hand, the glymphatic system, a clearance system that utilizes a unique system of perivascular channels to make the bridge between CSF and interstitial fluid in the brain and promote efficient elimination of soluble proteins and metabolites from the central nervous system (121), can be evoked, as it was proposed to participate in neuropropagation for viruses like chikungunya virus (122), SARS-CoV (41), and SARS-CoV-2 (94, 116, 123) and may account for the presence of the 3 viruses in CSF in our study. The capacity of viruses to infect ependymal cells surrounding the ventricles was also suggested to help dissemination toward the spinal cord (89, 90). Our data indicate that reference virus rOC/ATCC was the only HCoV-OC43 variant able to efficiently infect ependymal cells (Fig. 7) and produce large amounts of infectious particles in the spinal cord (Fig. 10) in immunocompetent mice. Our data support the hypothesis that infected ependymal cells serve to produce new infectious particles that will disseminate toward the spinal cord. The IFN-1 pathway has been shown to be cell type dependent for the murine coronavirus (MHV), as protection against infection seems to require the activity of different cell types altogether (66, 124) and this innate immunity-related pathway was also shown to efficiently abrogate infection of ependymal cells, diminish lethality, and reduce reovirus replication in the CNS (125). Like for the latter virus, increased infection of ependymal cells by our two S mutant viruses in IFNAR-KO mice strongly suggests that the efficient antiviral effect of IFN-1 depends on the status of the S protein cleavage. As shown for MHV (66) and other neurotropic viruses such as VSV (126), our data presage that the IFN-1 response would be of greater importance in specific cells (in our case, ependymal cells) in order to efficiently control viral spread and neuropathogenesis in immunocompetent mice in relation to the cleavage status of the S protein. As both mutant viruses almost completely lost their ability to efficiently infect ependymal cells in wild-type mice, these results presage that differential cleavage of the S protein may influence HCoV-OC43 tropism once the virus has already reached the brain and may partially help explain the slower and less efficient spread of the S mutant viruses in the whole CNS. The antiviral function of ependymal cells in the CNS was shown to be related to the IFN-1 pathway, as these cells constitutively express high levels of the IRF3 transcription factor (69). Furthermore, the interferon-stimulated gene (ISG) plasminogen activator inhibitor 1 (PAI-1), a member of the serine protein inhibitor (serpin) superfamily expressed in the CNS (127, 128), was shown to target host proteases, therefore modulating the cleavage of the influenza virus glycoprotein in response to IFN-1, thus reducing its infectivity (68). Our data presage that the same relation may occur between an efficient IFN-1 response and the cleavage of coronavirus S glycoprotein.
The observation that SARS-CoV-2 and other coronaviruses that infect humans are naturally neuroinvasive and neurotropic in both mice and humans (14, 15, 18, 20, 55, 94, 116, 129) underlines the need to further characterize viral and cellular determinants of these “neuroproperties.” Understanding mechanisms of CNS infection, from viral entry into neuronal cells to propagation to neighboring neurons, is essential to better conceive therapeutic strategies. IFN-1 response and coronavirus S protein cleavage have both been identified as potential targets to control infection in cell culture and in animal models before. However, the possible link between the two has still not been thoroughly studied. Our data point toward the importance of a differential cleavage of the S protein by diverse cellular proteases and presage that an adequate IFN-1 response participates by interfering with cell tropism and eventual dissemination within the CNS in relation to the S protein cleavage status. Here, our data suggest targeting both the IFN-1 response to ensure an effective immune response (especially in vulnerable populations such as the elderly and immunocompromised individuals) and cleavage of the S protein by host proteases at the same time. This would certainly constitute an interesting avenue to further investigate in order to identify more precise therapeutic targets to control viral dissemination in the specific context of CNS infection and potential short- and long-term neurovirulence of human coronaviruses.
MATERIALS AND METHODS
Ethics statement.
All animal experiments were approved by the Institutional Animal Care and Use Ethics Committee (IACUC) of the Institut national de la recherche scientifique (INRS) and conform to the Canadian Council on Animal Care (CCAC). Animal care and use protocol numbers 1304-02 and 1604-02 were issued by the IACUC of INRS for the animal experiments described herein.
Viruses and cell lines.
The wild-type reference virus HCoV-OC43 (VR-759) was obtained in 1980 from the American Type Culture Collection (ATCC). The recombinant HCoV-OC43 virus (rOC/ATCC) was generated using the full-length cDNA clone pBAC-OC43FL and displayed the same phenotypic properties as the wild-type virus, as previously described (77). The recombinant virus rOC/SG758R harbors a mutation in the gene coding for the spike glycoprotein of HCoV-OC43 at nucleotide (nt) 2272, corresponding to an amino acid change at position 758 (described elsewhere [55]). Both molecular clones pBAC-OC/SR903A (single mutation R903A; mutated at nucleotides 2707 and 2708) and pBAC-OC/SG758R-R903A (double mutant G758R and R903A; mutated at nucleotide 2272 and nucleotides 2707 and 2708) has been created using a recombineering approach with the en passant mutagenesis system described elsewhere (78). Each cDNA infectious clone was transfected into BHK-21 cells (ATCC CCL-10), amplified by two passages in the HRT-18 cell line, and sequenced to make sure that only the introduced G758R or R903A mutation was present and that no other mutations appeared. The HRT-18 cell line (a gift from the late David Brian, University of Tennessee) was cultured in minimal essential medium alpha (MEM-alpha; Life Technologies) supplemented with 10% (vol/vol) fetal bovine serum (FBS; PAA GE Healthcare) and was used to produce viral stocks. The LA-N-5 cell line (a kind gift of Stephan Ladisch, George Washington University School of Medicine) was cultured in RPMI medium supplemented with 15% (vol/vol) FBS, 10 mM HEPES, 1 mM sodium pyruvate, and 100 μM nonessential amino acids (Gibco-Invitrogen). LA-N-5 cells were differentiated into human neurons as previously described (130). Briefly, 1.25 × 104 LA-N-5 cells were seeded in RPMI medium supplemented with 20% (vol/vol) FBS, 10 mM HEPES, 1 mM sodium pyruvate, and 100 μM nonessential amino acids on 24-well glass coverslips previously coated with 0.1% gelatin for 2 h. The next day and every 2 days for 8 days, the medium was replaced with fresh RPMI medium supplemented with 10% (vol/vol) FBS, 10 mM HEPES, 1 mM sodium pyruvate, 100 μM nonessential amino acids, 50 μg/ml of gentamicin (Wisent), and 10 μM all-trans retinoic acid (Sigma-Aldrich). LA-N-5 cells were transduced with lentiviral vector particles expressing only the puromycin resistance gene (control; Origene; catalog no. PS100092V) or the puromycin resistance gene and human TPMRSS5/spinesin (Origene; catalog no. RC223774L3V), and stable populations and clones were selected with puromycin (Sigma-Aldrich; catalog no. P8833 at 1 μg/ml in complete RPMI medium). Total RNA was extracted with the RNeasy minikit (Qiagen) following the manufacturer’s instructions. The TMPRSS5/spinesin level of expression in different puromycin-selected populations and isolated clones of LA-N-5 cells was evaluated by RT-quantitative PCR (qRT-PCR) on the human TMPRSS5 gene (forward primer, 5′-GGCAGTGTGCTGGCGTTCCAT-3′, and reverse primer, 5′-CAGGGCCACGCTGGCCTGCCA-3′) and the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene (forward primer, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′, and reverse primer, 5′-AGCCTTCTCCATGGTGGTGAAGAC-3′).
Inhibitors.
The following inhibitors were used in this study: chloroquine phosphate [N4-(7-chloro-4-quinolinyl)-N1,N1-dimethyl-1,4-pentanediamine diphosphate salt; Sigma; catalog no. 50-63-5], MDL 28170 (EnzoBML; PI130), Z-FA-FMK (R&D Systems; FMKC01), and camostat mesylate (catalog no. 3193; Tocris Bioscience). The inhibitors were used at the concentrations indicated in the appropriate figure legends.
Protein extraction and Western blot analysis.
Proteins in the cell culture medium and cell-associated proteins were extracted using RIPA buffer (150 mM NaCl, 50 mM Tris [pH 7.4], 1% [vol/vol] NP-40, 0.25% [wt/vol] sodium deoxycholate, 1 mM EDTA) supplemented with the protease cocktail inhibitor (Sigma) and the Halt phosphatase inhibitor (Pierce). Harvested cells were pipetted up and down into RIPA buffer, incubated on ice for 20 min, and centrifuged for 10 min at 4°C and 17,000 × g, and supernatants were stored at −80°C until further analyzed. Protein concentrations were determined using the bicinchoninic acid (BCA) protein assay kit (Novagen) according to the manufacturer’s instructions. Equal amounts of proteins were subjected to SDS-PAGE using a Criterion 4 to 12% gradient gel or a Tris-glycine 4 to 15% gradient gel, transferred to a polyvinylidene difluoride (PVDF) membrane with a semidry Trans-Blot apparatus (Bio-Rad). Membranes were blocked overnight at 4°C with Tris-buffered saline (TBS) containing 1% (vol/vol) Tween (TBS-T) and 5% (wt/vol) nonfat milk and then incubated with monoclonal mouse anti-S protein antibody (1/2 of 4.3E4 hybridoma supernatant) for 1 h at room temperature. After three washes for 10 min with TBS-T, the membranes were incubated with a secondary anti-mouse antibody coupled to horseradish peroxidase (GE Life Sciences) and detection was performed using the enhanced chemiluminescence (ECL) kit (Bio-Rad) using Kodak-X-Omat L-S film (Kodak).
Neutralization of endosomal acidification.
Before infection, differentiated LA-N-5 cells in 24-well plates were pretreated with chloroquine phosphate at different concentrations indicated in the figure legends for 1 h at 37°C. The RPMI medium was removed and cells were infected at a defined multiplicity of infection (MOI) of 0.2 with reference and mutant viruses and incubated for 2 h at 37°C with chloroquine. Then, virus inocula were removed and fresh RPMI medium with chloroquine was added. At 16 hpi, cells were fixed 15 min in 4% paraformaldehyde (PFA). Immunofluorescence assay and cell count with the CellProfiler software were performed (https://cellprofiler.org/).
Protease inhibitor assay.
Before infection, differentiated LA-N-5 cells in 24-well plates were pretreated with MDL 28170, Z-FA-FMK, or camostat mesylate at different concentrations (indicated in figure legends) for 1 h at 37°C. The RPMI medium was removed and cells were infected at an MOI of 0.2 with reference and mutant viruses and incubated for 2 h at 37°C with inhibitors. Then, virus inocula were removed and fresh RMPI medium with chloroquine at 200 nM was added. At 16 hpi, cells were fixed for 15 min in 4% PFA. Immunofluorescence assay and cell count with the CellProfiler software were performed. Mixed primary cultures of mouse CNS, differentiated in 12-well plates, were pretreated with camostat mesylate at different concentrations for 1 h at 37°C. The neurobasal medium was removed and cells were infected at an MOI of 0.03 with reference and mutant viruses and incubated for 2 h at 37°C with inhibitors. The next step was identical to that for LA-N-5 cells.
Kinetics of HCoV-OC43 internalization assay.
To test virus internalization by the cells, HRT-18 and differentiated LA-N-5 cells in 24-well plates were incubated for 2 min with fresh cold RPMI medium. Then the medium was removed and viruses were incubated for 1 h on ice for virus binding. Ice-cold phosphate-buffered saline (PBS) was added to completely remove unbound viruses, and then fresh cold RPMI medium was added. Cells were then shifted to 37°C to allow internalization. After incubation for the desired time spans, cells were treated with chloroquine phosphate at 200 nM in order to prevent reinfection. At 16 hpi, cells were fixed with 4% PFA and an immunofluorescence assay (described below) was performed.
Mixed primary cultures of mouse CNS cells.
Embryos at 15 days of gestation were removed from pregnant anesthetized CD1 mice. The cortex and hippocampus of the embryonic pup brains (obtained from pregnant females; Charles River Canada) were harvested and placed in Hanks balanced salt solution (HBSS) medium, without Ca2+ and Mg2+, supplemented with 1.0 mM sodium pyruvate and 10 mM HEPES buffer. The tissues were incubated in 5 ml of HBSS plus trypsin-0.5% EDTA (ratio, 10:1) for 15 min at 37°C with gentle tilting to mix. After digestion, the tissues were washed for 5 min three times with HBSS, and the medium was removed and replaced with fresh HBSS medium (without Ca2+ and Mg2+, supplemented with 1.0 mM sodium pyruvate and 10 mM HEPES buffer). Tissues were gently pipetted up and down with a Pasteur pipette to dissociate the cells. After a decantation step of 5 min at room temperature, supernatants were transferred to a 50-ml tube with 36 ml of neurobasal medium (Invitrogen) supplemented with 0.5 mM GlutaMAX-I (Life Technologies), 10 mM HEPES buffer, B27 supplement (Life Technologies), gentamicin, and 10% (vol/vol) horse serum (Life Technologies). This step was performed twice to increase the final amount of cells. Cells were then seeded at 1 × 105/cm2 and grown on poly-d-lysine (50 μg/ml)-treated glass coverslips in the same medium, which was replaced with fresh neurobasal medium without horse medium the next day. The medium was changed every 2 days after, and the cultures were ready for infection after 7 days.
Infection of human cell lines and primary mouse CNS cultures.
The LA-N-5 cells and primary mouse neuronal cell cultures (PMNCs) were infected with recombinant HCoV-OC43 virus at the desired MOI for 2 h in neurobasal medium with B27-GlutaMAX-I (PMNCs) or RPMI medium supplemented with 1% (vol/vol) FBS (LA-N-5 cells). The inoculum was then discarded and replaced with fresh neurobasal medium with B27-GlutaMAX-I (PMNCs) or fresh RPMI medium supplemented with 2.5% (vol/vol) FBS (LA-N-5 cells) and incubated for different periods of time at 37°C.
Immunofluorescence on infected cells.
For immunofluorescence staining, HRT-18 cells, LA-N-5 cells, and primary murine CNS cell cultures were washed with sterile PBS and then fixed with 4% (wt/vol) paraformaldehyde for 30 min at room temperature. After washing, HRT-18 or LA-N-5 cells were permeabilized with 100% methanol at −20°C for 5 min and then incubated with primary antibody: a monoclonal mouse anti-S protein antibody (1/2 of 4.3.E4 hybridoma supernatant) for 1 h at room temperature. After three washes with PBS, cells were incubated in the dark for 1 h at room temperature with the secondary fluorescent antibody Alexa Fluor 488 goat anti-mouse (1/1,000; Life Technologies). Fixed and permeabilized primary murine cultures were incubated for 1 h at room temperature with a blocking solution (2% bovine serum albumin [BSA] in PBS supplemented with 0.1% Triton X-100) and then stained with primary antibodies: a polyclonal rabbit anti-S protein of bovine coronavirus (BCoV) at a 1/1,000 dilution and mouse monoclonal anti-microtubule-associated protein 2 (MAP2; at a dilution of 1/1,000) for 1 h at room temperature. After three washes with PBS, cells were incubated in the dark for 1 h at room temperature with the secondary fluorescent antibody Alexa Fluor 568 goat anti-rabbit (1/1,000; Life Technologies) or Alexa Fluor 488 anti-mouse (1/1,000; Life Technologies).
Virus propagation assay in cell cultures.
For propagation assays, cells were always seeded on glass coverslips following seeding procedures (see “Viruses and cell lines” above). Immediately after infection of LA-N-5 cells or murine primary cultures from the CNS (as described above), inocula were replaced with propagation media. Propagation medium was prepared as follows: 2× DMEM (Multicell) was supplemented with 2 mM GlutaMAX-I, 20 mM HEPES buffer, 100 μg/ml of gentamicin, 2 mM sodium pyruvate, and 200 μM nonessential amino acids. FBS at 5% (vol/vol) and 4% (vol/vol) B27 plus cobalamin at 0.013698 mg/ml were also respectively added for LA-N-5 cells and primary cultures. Supplemented 2× DMEM was then diluted 1/2 in either 4% (wt/vol) methylcellulose (semifluid medium) or H2O (fluid medium). Cells were overlaid with semifluid or fluid medium and incubated at 37°C for the desired time periods, after which supernatants were discarded and cells directly fixed with 4% PFA and stained as described above. Pictures were taken with a Zeiss LSM780 confocal microscope. Each sample was quantified (number of infected cells) using the cell image analysis software CellProfiler (https://cellprofiler.org/).
Mice, survival curves, body weight variations, and evaluation of clinical scores.
C57BL/6 mice (Charles River Laboratories) and IFNAR-KO mice (gift from Alain Lamarre, INRS-Institut Armand-Frappier) aged 22 days postnatal (dpn) or 10 dpn were inoculated respectively by the i.c. route with 101.5 50% tissue culture infective doses (TCID50) or the intranasal route with 103.5 TCID50 of recombinant virus, as previously described (38). Groups of 10 mice infected by each recombinant virus were observed on a daily basis over a period of 21 dpi, and survival and weight variations were evaluated. Clinical scores were evaluated using a scale with 5 distinctive levels (0 to 4), where 0 was used for the asymptomatic mouse, 1 for mice with early hunched back, 2 for mice presenting slight social isolation, weight loss, and abnormal gait, 3 for mice presenting total social isolation, ruffled fur, hunched back, weight loss, and almost no movement; and 4 for mice that were in a moribund state or dead (55).
Evaluation of neuroinvasiveness.
Ten-day-old mice were subjected to intranasal (5 μl/nostril) inoculation of rOC/ATCC, rOC/SG758R, or rOC/SR903A using 103.5 TCID50 per 10 μl. Sham-infected mice received PBS. Different regions of the brain (Fig. 6) from peritoneally anesthetized mice (ketamine at 200 mg/kg of body weight and xylazine at 10 mg/kg) were harvested at 2, 3, and 4 dpi and frozen at −80°C until further analysis. Tissue was shredded/lysed by extensive agitation in 1 ml of QIAzol lysis reagent (Qiagen) supplemented with shredding beads, and total RNA was extracted by the QIAzol/chloroform/propanol manufacturer’s procedure, dosed using a NanoDrop 1000 spectrophotometer (NanoDrop), and frozen at −80°C. Virus RNA copy numbers were quantified in triplicate by real-time RT-PCR using the TaqMan RNA-to-CT 1-step kit (Applied Biosystems/Life Technologies) in a 20-μl reaction mixture with 10 μl of 2× TaqMan RT-PCR mix (containing ROX [carboxyrhodamine] as a passive reference dye), 900 nM forward and reverse primers targeting a 68-bp region of the HCoV-OC43 M gene (forward primer OC43-FP, 5′-ATGTTAGGCCGATAATTGAGGACTAT-3′, nt 433 to 458, and reverse primer OC43-RP, 5′-AATGTAAAGATGGCCGCGTATT-3′, nt 479 to 500), 200 nM 6-carboxyfluorescein (FAM) BHQ1-TP probe (OC43-TP, FAM-5′-CATACTCTGACGGTCACAAT-3′, nt 459 to 478), 0.5 μl of 40× TaqMan RT enzyme mix, and 800 ng of extracted brain RNA. Serially diluted cRNA standards were used for the generation of a standard curve. Amplification and detection were performed in a StepOnePlus real-time PCR system apparatus and analyzed with StepOne software version 2.3 (Applied Biosystems). The limit of detection was defined as the average signal obtained in corresponding negative controls for each organ.
Viral RNA extraction from mouse tissue after intracranial infection.
Brain, spinal cord, and CSF were collected as previously described (131). Viral RNA was then extracted as described above, and quantification by TaqMan qRT-PCR was performed.
Evaluation of infectious virus production.
Mouse brain and spinal cord or cell culture supernatants were processed for the presence and quantification of infectious virus by an indirect immunoperoxidase assay (IPA) on HRT-18 cells, as previously described (132). Briefly, HRT-18 cells were incubated with the mouse primary antibody 4.3E4 (dilution, 1/50), which detects the S protein of HCoV-OC43. After three PBS washes, cells were incubated with a secondary horseradish peroxidase-conjugated goat anti-mouse immunoglobulin antibody diluted 1/500 (Kierkegaard & Perry Laboratories). Finally, immune complexes were detected by incubation with 0.025% (wt/vol) 3,3-diaminobenzidine tetrahydrochloride (Sigma-Aldrich) and 0.01% (vol/vol) hydrogen peroxide in 1× PBS, and infectious virus titers were calculated by the Karber method, as previously described (132).
Immunofluorescence on CNS sections.
Confocal immunofluorescence was performed either after intracranial infection of 22-day-old mice by either rOC/ATCC, rOC/SG758R, or rOC/SR903A, for which perfusion with 4% (wt/vol) PFA was performed every 2 days between 3 and 7 days postinfection, or after intranasal infection on 10-day-old mice, for which perfusion was performed every day for 5 days. For intracranial infection, brain sections were sagittally sliced in 50-μm-thick sections with a cryostat (HM 525; Microm), and for intranasal infection, heads were harvested and fur, skin, and lower jaw were removed. Whole heads were decalcified in 6% EDTA (pH 8) at 4°C for 7 days then transferred to a 30% sucrose solution before being processed 3 days later. Serial sections were collected in PBS, treated for 10 min with H2O2 to disrupt erythrocytes, washed twice for 5 min in PBS, and permeabilized for 2 h in PBS supplemented with 0.1% (vol/vol) Triton X-100. Sections were further incubated for 1 h in 0.1% Triton X-100–PBS supplemented 1% (vol/vol) horse serum and then overnight at 4°C in 0.05% Triton X-100–PBS containing 1% horse serum and BCoV anti-S (1/500) (55). Sections were washed 3 times (15 min each) in 0.05% Triton X-100–PBS and incubated for 2 h in 0.05% Triton X-100–PBS supplemented with 1% horse serum and 1/500 adequate Alexa Fluor 488-coupled secondary antibodies (Life Technologies). Immunostained sections were washed twice (5 min each) in 0.05% Triton X-100–PBS, counterstained for nuclei with 10 μg/ml of 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen), and then washed again 4 times (15 min each) in 0.05% Triton X-100–PBS. Sections were then mounted in ProLong Diamond antifade mounting medium (Molecular Probes) on glass slides and imaged on a Zeiss LSM780 confocal microscope. All immunostaining steps were carried out at room temperature with agitation unless otherwise stipulated.
Synthetic peptides and in vitro cleavage assay.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed for quantification of the N-321, N-322, S2-ref, and S2-mutant peptides. The Michaelis-Menten constants (Km and Vmax) of cathepsin B (catalog no. 953-CY), cathepsin L (catalog no. 952-CY), and spinesin (catalog no. 2495-SE-010) were determined in a 30-min kinetic enzymatic assay with the recommended specifications provided by the manufacturer with fluorogenic substrate (catalog no. ES008 and ES014; all reagents were from R&D Systems). For furin, kindly provided by Robert Day, Km and Vmax were previously determined (133). Instrumentation, experimental conditions, and software analysis were also previously described (133). Each enzyme was incubated for 30 min with peptide N-321 (VDYSKNRRSRRAITTGY) (sequences in bold are the recognition sites for the proteases, and underlining indicates the specific amino acids where the protease cuts), N-322 (VDYSKNRRSR*GAITTGY), S2-ref (GCLGSECSKASSRSAIEDL), or S2-mutant (GCLGSECSKASS*ASAIEDL), at their respective Km values. In vitro cleavage by recombinant enzymes was performed and analyzed by LC-MS/MS as previously described (134).
Statistical tests.
For cell experiments, statistical analyses were conducted by one-way analysis of variance (ANOVA), followed by Tukey’s post hoc test, or a t test. For mouse experiments, results were compared using Kruskal-Wallis and Mann-Whitney nonparametric tests. Survival rates were plotted as Kaplan-Meier survival curves and compared using the log rank (Mantel-Cox) test. Statistical significance was defined as a P value of <0.05.
ACKNOWLEDGMENTS
We thank J. Tremblay for excellent technical assistance and M. Dubé and J. K. Millet for excellent advice and critical reviews of the manuscript.
This work was supported by grant no. MT-9203 from the CIHR to Pierre J. Talbot.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Talbot PJ, Jacomy HD, Desforges M. 2008. Pathogenesis of human coronaviruses other than severe acute respiratory syndrome coronavirus, p 313–324. In Perlman S, Gallagher T, Snijder EJ (ed), Nidoviruses. ASM Press, Washington, DC. [Google Scholar]
- 2.Hamre D, Procknow JJ. 1966. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med 121:190–193. 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 3.McIntosh K, Becker WB, Chanock RM. 1967. Growth in suckling-mouse brain of “IBV-like” viruses from patients with upper respiratory tract disease. Proc Natl Acad Sci U S A 58:2268–2273. 10.1073/pnas.58.6.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, Wertheim-van Dillen PM, Kaandorp J, Spaargaren J, Berkhout B. 2004. Identification of a new human coronavirus. Nat Med 10:368–373. 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, Wong BH, Poon RW, Cai JJ, Luk WK, Poon LL, Wong SS, Guan Y, Peiris JS, Yuen KY. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol 79:884–895. 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967–1976. 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 7.Fouchier RA, Kuiken T, Schutten M, van Amerongen G, van Doornum GJ, van den Hoogen BG, Peiris M, Lim W, Stohr K, Osterhaus AD. 2003. Aetiology: Koch’s postulates fulfilled for SARS virus. Nature 423:240. 10.1038/423240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ, SARS Working Group . 2003. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med 348:1953–1966. 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 9.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 10.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, Bleicker T, Brunink S, Schneider J, Schmidt ML, Mulders DG, Haagmans BL, van der Veer B, van den Brink S, Wijsman L, Goderski G, Romette JL, Ellis J, Zambon M, Peiris M, Goossens H, Reusken C, Koopmans MP, Drosten C. 2020. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill 25:2000045. 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. 2020. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vabret A, Dina J, Brison E, Brouard J, Freymuth F. 2009. Human coronaviruses. Pathol Biol (Paris) 57:149–160. 10.1016/j.patbio.2008.02.018. (In French.) 10.1016/j.patbio.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desforges M, Le Coupanec A, Dubeau P, Bourgouin A, Lajoie L, Dube M, Talbot PJ. 2019. Human coronaviruses and other respiratory viruses: underestimated opportunistic pathogens of the central nervous system? Viruses 12:14. 10.3390/v12010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arbour N, Day R, Newcombe J, Talbot PJ. 2000. Neuroinvasion by human respiratory coronaviruses. J Virol 74:8913–8921. 10.1128/jvi.74.19.8913-8921.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desforges M, Le Coupanec A, Stodola JK, Meessen-Pinard M, Talbot PJ. 2014. Human coronaviruses: viral and cellular factors involved in neuroinvasiveness and neuropathogenesis. Virus Res 194:145–158. 10.1016/j.virusres.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gu J, Gong E, Zhang B, Zheng J, Gao Z, Zhong Y, Zou W, Zhan J, Wang S, Xie Z, Zhuang H, Wu B, Zhong H, Shao H, Fang W, Gao D, Pei F, Li X, He Z, Xu D, Shi X, Anderson VM, Leong AS. 2005. Multiple organ infection and the pathogenesis of SARS. J Exp Med 202:415–424. 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song E, Zhang C, Israelow B, Lu-Culligan A, Prado AV, Skriabine S, Lu P, Weizman OE, Liu F, Dai Y, Szigeti-Buck K, Yasumoto Y, Wang G, Castaldi C, Heltke J, Ng E, Wheeler J, Alfajaro MM, Levavasseur E, Fontes B, Ravindra NG, Van Dijk D, Mane S, Gunel M, Ring A, Kazmi SAJ, Zhang K, Wilen CB, Horvath TL, Plu I, Haik S, Thomas JL, Louvi A, Farhadian SF, Huttner A, Seilhean D, Renier N, Bilguvar K, Iwasaki A. 2021. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J Exp Med 218:e20202135. 10.1084/jem.20202135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arabi YM, Harthi A, Hussein J, Bouchama A, Johani S, Hajeer AH, Saeed BT, Wahbi A, Saedy A, AlDabbagh T, Okaili R, Sadat M, Balkhy H. 2015. Severe neurologic syndrome associated with Middle East respiratory syndrome corona virus (MERS-CoV). Infection 43:495–501. 10.1007/s15010-015-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeh EA, Collins A, Cohen ME, Duffner PK, Faden H. 2004. Detection of coronavirus in the central nervous system of a child with acute disseminated encephalomyelitis. Pediatrics 113:e73–e76. 10.1542/peds.113.1.e73. [DOI] [PubMed] [Google Scholar]
- 21.Algahtani H, Subahi A, Shirah B. 2016. Neurological complications of Middle East respiratory syndrome coronavirus: a report of two cases and review of the literature. Case Rep Neurol Med 2016:3502683. 10.1155/2016/3502683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau KK, Yu WC, Chu CM, Lau ST, Sheng B, Yuen KY. 2004. Possible central nervous system infection by SARS coronavirus. Emerg Infect Dis 10:342–344. 10.3201/eid1002.030638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turgay C, Emine T, Ozlem K, Muhammet SP, Haydar AT. 2015. A rare cause of acute flaccid paralysis: human coronaviruses. J Pediatr Neurosci 10:280–281. 10.4103/1817-1745.165716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y, Xu X, Chen Z, Duan J, Hashimoto K, Yang L, Liu C, Yang C. 2020. Nervous system involvement after infection with COVID-19 and other coronaviruses. Brain Behav Immun 10.1016/j.bbi.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li YC, Bai WZ, Hashikawa T. 2020. Response to commentary on “The neuroinvasive potential of SARS-CoV-2 may play a role in the respiratory failure of COVID-19 patients.” J Med Virol 92:707–709. 10.1002/jmv.25824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li YC, Bai WZ, Hashikawa T. 2020. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J Med Virol 92:552–555. 10.1002/jmv.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baig AM, Khaleeq A, Ali U, Syeda H. 2020. Evidence of the COVID-19 virus targeting the CNS: tissue distribution, host-virus interaction, and proposed neurotropic mechanisms. ACS Chem Neurosci 11:995–998. 10.1021/acschemneuro.0c00122. [DOI] [PubMed] [Google Scholar]
- 28.Pleasure SJ, Green AJ, Josephson SA. 2020. The spectrum of neurologic disease in the severe acute respiratory syndrome coronavirus 2 pandemic infection: neurologists move to the frontlines. JAMA Neurol 77:679. 10.1001/jamaneurol.2020.1065. [DOI] [PubMed] [Google Scholar]
- 29.Mao L, Jin H, Wang M, Hu Y, Chen S, He Q, Chang J, Hong C, Zhou Y, Wang D, Miao X, Li Y, Hu B. 2020. Neurologic manifestations of hospitalized patients with coronavirus disease 2019 in Wuhan, China. JAMA Neurol 77:683. 10.1001/jamaneurol.2020.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lechien JR, Chiesa-Estomba CM, De Siati DR, Horoi M, Le Bon SD, Rodriguez A, Dequanter D, Blecic S, El Afia F, Distinguin L, Chekkoury-Idrissi Y, Hans S, Delgado IL, Calvo-Henriquez C, Lavigne P, Falanga C, Barillari MR, Cammaroto G, Khalife M, Leich P, Souchay C, Rossi C, Journe F, Hsieh J, Edjlali M, Carlier R, Ris L, Lovato A, De Filippis C, Coppee F, Fakhry N, Ayad T, Saussez S. 2020. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): a multicenter European study. Eur Arch Otorhinolaryngol 277:2251–2261. 10.1007/s00405-020-05965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou L, Zhang M, Wang J, Gao J. 2020. Sars-Cov-2: underestimated damage to nervous system. Travel Med Infect Dis 36:101642. 10.1016/j.tmaid.2020.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poyiadji N, Shahin G, Noujaim D, Stone M, Patel S, Griffith B. 2020. COVID-19-associated acute hemorrhagic necrotizing encephalopathy: CT and MRI features. Radiology 296:E119–E120. 10.1148/radiol.2020201187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zanin L, Saraceno G, Panciani PP, Renisi G, Signorini L, Migliorati K, Fontanella MM. 2020. SARS-CoV-2 can induce brain and spine demyelinating lesions. Acta Neurochir (Wien) 162:1491–1494. 10.1007/s00701-020-04374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coen M, Jeanson G, Culebras Almeida LA, Hubers A, Stierlin F, Najjar I, Ongaro M, Moulin K, Makrygianni M, Leemann B, Kronig I, Bertrand J, Reny JL, Schibler M, Serratrice J. 2020. Guillain-Barre syndrome as a complication of SARS-CoV-2 infection. Brain Behav Immun 87:111–112. 10.1016/j.bbi.2020.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alberti P, Beretta S, Piatti M, Karantzoulis A, Piatti ML, Santoro P, Vigano M, Giovannelli G, Pirro F, Montisano DA, Appollonio I, Ferrarese C. 2020. Guillain-Barre syndrome related to COVID-19 infection. Neurol Neuroimmunol Neuroinflamm 7:e741. 10.1212/NXI.0000000000000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vonck K, Garrez I, De Herdt V, Hemelsoet D, Laureys G, Raedt R, Boon P. 2020. Neurological manifestations and neuro-invasive mechanisms of the severe acute respiratory syndrome coronavirus type 2. Eur J Neurol 27:1578–1587. 10.1111/ene.14329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Felice FG, Tovar-Moll F, Moll J, Munoz DP, Ferreira ST. 2020. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the central nervous system. Trends Neurosci 43:355–357. 10.1016/j.tins.2020.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacomy H, St-Jean JR, Brison E, Marceau G, Desforges M, Talbot PJ. 2010. Mutations in the spike glycoprotein of human coronavirus OC43 modulate disease in BALB/c mice from encephalitis to flaccid paralysis and demyelination. J Neurovirol 16:279–293. 10.3109/13550284.2010.497806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacomy H, Talbot PJ. 2003. Vacuolating encephalitis in mice infected by human coronavirus OC43. Virology 315:20–33. 10.1016/s0042-6822(03)00323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Butler N, Pewe L, Trandem K, Perlman S. 2006. Murine encephalitis caused by HCoV-OC43, a human coronavirus with broad species specificity, is partly immune-mediated. Virology 347:410–421. 10.1016/j.virol.2005.11.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Netland J, Meyerholz DK, Moore S, Cassell M, Perlman S. 2008. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J Virol 82:7264–7275. 10.1128/JVI.00737-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morfopoulou S, Brown JR, Davies EG, Anderson G, Virasami A, Qasim W, Chong WK, Hubank M, Plagnol V, Desforges M, Jacques TS, Talbot PJ, Breuer J. 2016. Human coronavirus OC43 associated with fatal encephalitis. N Engl J Med 375:497–498. 10.1056/NEJMc1509458. [DOI] [PubMed] [Google Scholar]
- 43.Nilsson A, Edner N, Albert J, Ternhag A. 2020. Fatal encephalitis associated with coronavirus OC43 in an immunocompromised child. Infect Dis (Lond) 52:419–422. 10.1080/23744235.2020.1729403. [DOI] [PubMed] [Google Scholar]
- 44.Moriguchi T, Harii N, Goto J, Harada D, Sugawara H, Takamino J, Ueno M, Sakata H, Kondo K, Myose N, Nakao A, Takeda M, Haro H, Inoue O, Suzuki-Inoue K, Kubokawa K, Ogihara S, Sasaki T, Kinouchi H, Kojin H, Ito M, Onishi H, Shimizu T, Sasaki Y, Enomoto N, Ishihara H, Furuya S, Yamamoto T, Shimada S. 2020. A first case of meningitis/encephalitis associated with SARS-coronavirus-2. Int J Infect Dis 94:55–58. 10.1016/j.ijid.2020.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao P, Huang Y, Wu Z, Sun H, Ma W, Fang T. 2020. A case of coronavirus HKU1 encephalitis. Acta Virol 64:261–263. 10.4149/av_2020_214. [DOI] [PubMed] [Google Scholar]
- 46.Masters P. 2013. Coronaviridae, p 825–858. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed, vol 1. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 47.Vlasak R, Luytjes W, Spaan W, Palese P. 1988. Human and bovine coronaviruses recognize sialic acid-containing receptors similar to those of influenza C viruses. Proc Natl Acad Sci U S A 85:4526–4529. 10.1073/pnas.85.12.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol 77:8801–8811. 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gallagher TM, Buchmeier MJ. 2001. Coronavirus spike proteins in viral entry and pathogenesis. Virology 279:371–374. 10.1006/viro.2000.0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klenk HD, Garten W. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol 2:39–43. 10.1016/0966-842x(94)90123-6. [DOI] [PubMed] [Google Scholar]
- 51.Millet JK, Whittaker GR. 2015. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res 202:120–134. 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Haan CA, Stadler K, Godeke GJ, Bosch BJ, Rottier PJ. 2004. Cleavage inhibition of the murine coronavirus spike protein by a furin-like enzyme affects cell-cell but not virus-cell fusion. J Virol 78:6048–6054. 10.1128/JVI.78.11.6048-6054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Follis KE, York J, Nunberg JH. 2006. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology 350:358–369. 10.1016/j.virol.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simmons G, Bertram S, Glowacka I, Steffen I, Chaipan C, Agudelo J, Lu K, Rennekamp AJ, Hofmann H, Bates P, Pohlmann S. 2011. Different host cell proteases activate the SARS-coronavirus spike-protein for cell-cell and virus-cell fusion. Virology 413:265–274. 10.1016/j.virol.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le Coupanec A, Desforges M, Meessen-Pinard M, Dube M, Day R, Seidah NG, Talbot PJ. 2015. Cleavage of a neuroinvasive human respiratory virus spike glycoprotein by proprotein convertases modulates neurovirulence and virus spread within the central nervous system. PLoS Pathog 11:e1005261. 10.1371/journal.ppat.1005261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH, Nitsche A, Muller MA, Drosten C, Pohlmann S. 2020. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271–280.e8. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. 2020. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 11:1620. 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF. 2020. The proximal origin of SARS-CoV-2. Nat Med 26:450–452. 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reinke LM, Spiegel M, Plegge T, Hartleib A, Nehlmeier I, Gierer S, Hoffmann M, Hofmann-Winkler H, Winkler M, Pohlmann S. 2017. Different residues in the SARS-CoV spike protein determine cleavage and activation by the host cell protease TMPRSS2. PLoS One 12:e0179177. 10.1371/journal.pone.0179177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Belouzard S, Millet JK, Licitra BN, Whittaker GR. 2012. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4:1011–1033. 10.3390/v4061011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Belouzard S, Chu VC, Whittaker GR. 2009. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A 106:5871–5876. 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamada Y, Liu DX. 2009. Proteolytic activation of the spike protein at a novel RRRR/S motif is implicated in furin-dependent entry, syncytium formation, and infectivity of coronavirus infectious bronchitis virus in cultured cells. J Virol 83:8744–8758. 10.1128/JVI.00613-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lai AL, Li Y, Tamm LK. 2005. Interplay of proteins and lipids in virus entry by membrane fusion, p 279–303. Tamm LK (ed), Protein-lipid interactions. Wiley-VCH, Weinheim, Germany. [Google Scholar]
- 64.St-Jean JR, Desforges M, Talbot PJ. 2006. Genetic evolution of human coronavirus OC43 in neural cell culture. Adv Exp Med Biol 581:499–502. 10.1007/978-0-387-33012-9_88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cheng Y, Skinner DD, Lane TE. 2019. Innate immune responses and viral-induced neurologic disease. J Clin Med 8:3. 10.3390/jcm8010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rose KM, Weiss SR. 2009. Murine coronavirus cell type dependent interaction with the type I interferon response. Viruses 1:689–712. 10.3390/v1030689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Foronjy RF, Taggart CC, Dabo AJ, Weldon S, Cummins N, Geraghty P. 2015. Type-I interferons induce lung protease responses following respiratory syncytial virus infection via RIG-I-like receptors. Mucosal Immunol 8:161–175. 10.1038/mi.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dittmann M, Hoffmann HH, Scull MA, Gilmore RH, Bell KL, Ciancanelli M, Wilson SJ, Crotta S, Yu Y, Flatley B, Xiao JW, Casanova JL, Wack A, Bieniasz PD, Rice CM. 2015. A serpin shapes the extracellular environment to prevent influenza A virus maturation. Cell 160:631–643. 10.1016/j.cell.2015.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tarassishin L, Bauman A, Suh HS, Lee SC. 2013. Anti-viral and anti-inflammatory mechanisms of the innate immune transcription factor interferon regulatory factor 3: relevance to human CNS diseases. J Neuroimmune Pharmacol 8:132–144. 10.1007/s11481-012-9360-5. [DOI] [PubMed] [Google Scholar]
- 70.Zang R, Gomez Castro MF, McCune BT, Zeng Q, Rothlauf PW, Sonnek NM, Liu Z, Brulois KF, Wang X, Greenberg HB, Diamond MS, Ciorba MA, Whelan SPJ, Ding S. 2020. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol 5:eabc3582. 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. 2019. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after coronavirus infection. J Virol 93:e01815-18. 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lau SY, Wang P, Mok BW, Zhang AJ, Chu H, Lee AC, Deng S, Chen P, Chan KH, Song W, Chen Z, To KK, Chan JF, Yuen KY, Chen H. 2020. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerg Microbes Infect 9:837–842. 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Andre NM, Cossic B, Davies E, Miller AD, Whittaker GR. 2019. Distinct mutation in the feline coronavirus spike protein cleavage activation site in a cat with feline infectious peritonitis-associated meningoencephalomyelitis. JFMS Open Rep 5:2055116919856103. 10.1177/2055116919856103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Glowacka I, Bertram S, Muller MA, Allen P, Soilleux E, Pfefferle S, Steffen I, Tsegaye TS, He Y, Gnirss K, Niemeyer D, Schneider H, Drosten C, Pohlmann S. 2011. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol 85:4122–4134. 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Millet JK, Whittaker GR. 2014. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci U S A 111:15214–15219. 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bertram S, Dijkman R, Habjan M, Heurich A, Gierer S, Glowacka I, Welsch K, Winkler M, Schneider H, Hofmann-Winkler H, Thiel V, Pohlmann S. 2013. TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium. J Virol 87:6150–6160. 10.1128/JVI.03372-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.St-Jean JR, Desforges M, Almazan F, Jacomy H, Enjuanes L, Talbot PJ. 2006. Recovery of a neurovirulent human coronavirus OC43 from an infectious cDNA clone. J Virol 80:3670–3674. 10.1128/JVI.80.7.3670-3674.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol Biol 634:421–430. 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 79.Jaimes JA, Millet JK, Whittaker GR. 2020. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. iScience 23:101212. 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jaimes JA, Andre NM, Chappie JS, Millet JK, Whittaker GR. 2020. Phylogenetic analysis and structural modeling of SARS-CoV-2 spike protein reveals an evolutionary distinct and proteolytically sensitive activation loop. J Mol Biol 432:3309–3325. 10.1016/j.jmb.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qiu Z, Hingley ST, Simmons G, Yu C, Das SJ, Bates P, Weiss SR. 2006. Endosomal proteolysis by cathepsins is necessary for murine coronavirus mouse hepatitis virus type 2 spike-mediated entry. J Virol 80:5768–5776. 10.1128/JVI.00442-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shirato K, Kanou K, Kawase M, Matsuyama S. 2017. Clinical isolates of human coronavirus 229E bypass the endosome for cell entry. J Virol 91:e01387-16. 10.1128/JVI.01387-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Simmons G, Reeves JD, Rennekamp AJ, Amberg SM, Piefer AJ, Bates P. 2004. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci U S A 101:4240–4245. 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Simmons G, Gosalia DN, Rennekamp AJ, Reeves JD, Diamond SL, Bates P. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc Natl Acad Sci U S A 102:11876–11881. 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gierer S, Bertram S, Kaup F, Wrensch F, Heurich A, Kramer-Kuhl A, Welsch K, Winkler M, Meyer B, Drosten C, Dittmer U, von Hahn T, Simmons G, Hofmann H, Pohlmann S. 2013. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol 87:5502–5511. 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shirato K, Kawase M, Matsuyama S. 2018. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology 517:9–15. 10.1016/j.virol.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nash TC, Buchmeier MJ. 1997. Entry of mouse hepatitis virus into cells by endosomal and nonendosomal pathways. Virology 233:1–8. 10.1006/viro.1997.8609. [DOI] [PubMed] [Google Scholar]
- 88.Dube M, Le Coupanec A, Wong AHM, Rini JM, Desforges M, Talbot PJ. 2018. Axonal transport enables neuron-to-neuron propagation of human coronavirus OC43. J Virol 92:e00404-18. 10.1128/JVI.00404-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Plakhov IV, Arlund EE, Aoki C, Reiss CS. 1995. The earliest events in vesicular stomatitis virus infection of the murine olfactory neuroepithelium and entry of the central nervous system. Virology 209:257–262. 10.1006/viro.1995.1252. [DOI] [PubMed] [Google Scholar]
- 90.Tuomanen E. 1996. Entry of pathogens into the central nervous system. FEMS Microbiol Rev 18:289–299. 10.1111/j.1574-6976.1996.tb00245.x. [DOI] [PubMed] [Google Scholar]
- 91.Meessen-Pinard M, Le Coupanec A, Desforges M, Talbot PJ. 2017. Pivotal role of receptor-interacting protein kinase 1 and mixed lineage kinase domain-like in neuronal cell death induced by the human neuroinvasive coronavirus OC43. J Virol 91:e01513-16. 10.1128/JVI.01513-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jacomy H, Fragoso G, Almazan G, Mushynski WE, Talbot PJ. 2006. Human coronavirus OC43 infection induces chronic encephalitis leading to disabilities in BALB/C mice. Virology 349:335–346. 10.1016/j.virol.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bilinska K, Jakubowska P, Von Bartheld CS, Butowt R. 2020. Expression of the SARS-CoV-2 entry proteins, ACE2 and TMPRSS2, in cells of the olfactory epithelium: identification of cell types and trends with age. ACS Chem Neurosci 11:1555–1562. 10.1021/acschemneuro.0c00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Butowt R, Bilinska K. 2020. SARS-CoV-2: olfaction, brain infection, and the urgent need for clinical samples allowing earlier virus detection. ACS Chem Neurosci 11:1200–1203. 10.1021/acschemneuro.0c00172. [DOI] [PubMed] [Google Scholar]
- 95.Leparc-Goffart I, Hingley ST, Chua MM, Jiang X, Lavi E, Weiss SR. 1997. Altered pathogenesis of a mutant of the murine coronavirus MHV-A59 is associated with a Q159L amino acid substitution in the spike protein. Virology 239:1–10. 10.1006/viro.1997.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Regan AD, Shraybman R, Cohen RD, Whittaker GR. 2008. Differential role for low pH and cathepsin-mediated cleavage of the viral spike protein during entry of serotype II feline coronaviruses. Vet Microbiol 132:235–248. 10.1016/j.vetmic.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Luytjes W, Sturman LS, Bredenbeek PJ, Charite J, van der Zeijst BA, Horzinek MC, Spaan WJ. 1987. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology 161:479–487. 10.1016/0042-6822(87)90142-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thomas G. 2002. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol 3:753–766. 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shirato K, Kawase M, Matsuyama S. 2013. Middle East respiratory syndrome coronavirus infection mediated by the transmembrane serine protease TMPRSS2. J Virol 87:12552–12561. 10.1128/JVI.01890-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huang IC, Bosch BJ, Li F, Li W, Lee KH, Ghiran S, Vasilieva N, Dermody TS, Harrison SC, Dormitzer PR, Farzan M, Rottier PJ, Choe H. 2006. SARS coronavirus, but not human coronavirus NL63, utilizes cathepsin L to infect ACE2-expressing cells. J Biol Chem 281:3198–3203. 10.1074/jbc.M508381200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kawase M, Shirato K, van der Hoek L, Taguchi F, Matsuyama S. 2012. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J Virol 86:6537–6545. 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Qian Z, Dominguez SR, Holmes KV. 2013. Role of the spike glycoprotein of human Middle East respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS One 8:e76469. 10.1371/journal.pone.0076469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Burkard C, Verheije MH, Wicht O, van Kasteren SI, van Kuppeveld FJ, Haagmans BL, Pelkmans L, Rottier PJ, Bosch BJ, de Haan CA. 2014. Coronavirus cell entry occurs through the endo-/lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog 10:e1004502. 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Matsuyama S, Nagata N, Shirato K, Kawase M, Takeda M, Taguchi F. 2010. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol 84:12658–12664. 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shulla A, Heald-Sargent T, Subramanya G, Zhao J, Perlman S, Gallagher T. 2011. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol 85:873–882. 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Phillips JM, Gallagher T, Weiss SR. 2017. Neurovirulent murine coronavirus JHM.SD uses cellular zinc metalloproteases for virus entry and cell-cell fusion. J Virol 91:e01564-16. 10.1128/JVI.01564-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Heald-Sargent T, Gallagher T. 2012. Ready, set, fuse! The coronavirus spike protein and acquisition of fusion competence. Viruses 4:557–580. 10.3390/v4040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Seidah NG, Prat A. 2012. The biology and therapeutic targeting of the proprotein convertases. Nat Rev Drug Discov 11:367–383. 10.1038/nrd3699. [DOI] [PubMed] [Google Scholar]
- 109.Millet JK, Goldstein ME, Labitt RN, Hsu HL, Daniel S, Whittaker GR. 2016. A camel-derived MERS-CoV with a variant spike protein cleavage site and distinct fusion activation properties. Emerg Microbes Infect 5:e126. 10.1038/emi.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shen LW, Mao HJ, Wu YL, Tanaka Y, Zhang W. 2017. TMPRSS2: a potential target for treatment of influenza virus and coronavirus infections. Biochimie 142:1–10. 10.1016/j.biochi.2017.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qiu S, Leung A, Bo Y, Kozak RA, Anand SP, Warkentin C, Salambanga FDR, Cui J, Kobinger G, Kobasa D, Cote M. 2018. Ebola virus requires phosphatidylinositol (3,5) bisphosphate production for efficient viral entry. Virology 513:17–28. 10.1016/j.virol.2017.09.028. [DOI] [PubMed] [Google Scholar]
- 112.Millet JK, Tang T, Nathan L, Jaimes JA, Hsu HL, Daniel S, Whittaker GR. 2019. Production of pseudotyped particles to study highly pathogenic coronaviruses in a biosafety level 2 setting. J Vis Exp 2019(145):e59010. 10.3791/59010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Li W, Wicht O, van Kuppeveld FJM, He Q, Rottier PJ, Bosch B-J. 2015. A single point mutation creating a furin cleavage site in the spike protein renders porcine epidemic diarrhea coronavirus trypsin independent for cell entry and fusion. J Virol 89:8077–8081. 10.1128/JVI.00356-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shirato K, Matsuyama S, Ujike M, Taguchi F. 2011. Role of proteases in the release of porcine epidemic diarrhea virus from infected cells. J Virol 85:7872–7880. 10.1128/JVI.00464-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Desforges M, Le Coupanec A, Brison E, Meessen-Pinard M, Talbot PJ. 2014. Neuroinvasive and neurotropic human respiratory coronaviruses: potential neurovirulent agents in humans. Adv Exp Med Biol 807:75–96. 10.1007/978-81-322-1777-0_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Briguglio M, Bona A, Porta M, Dell'Osso B, Pregliasco FE, Banfi G. 2020. Disentangling the hypothesis of host dysosmia and SARS-CoV-2: the bait symptom that hides neglected neurophysiological routes. Front Physiol 11:671. 10.3389/fphys.2020.00671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.DosSantos MF, Devalle S, Aran V, Capra D, Roque NR, Coelho-Aguiar JM, Spohr T, Subilhaga JG, Pereira CM, D’Andrea Meira I, Niemeyer Soares Filho P, Moura-Neto V. 2020. Neuromechanisms of SARS-CoV-2: a review. Front Neuroanat 14:37. 10.3389/fnana.2020.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Andriuta D, Roger PA, Thibault W, Toublanc B, Sauzay C, Castelain S, Godefroy O, Brochot E. 2020. COVID-19 encephalopathy: detection of antibodies against SARS-CoV-2 in CSF. J Neurol 267:2810–2811. 10.1007/s00415-020-09975-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Domingues RB, Mendes-Correa MC, de Moura Leite FBV, Sabino EC, Salarini DZ, Claro I, Santos DW, de Jesus JG, Ferreira NE, Romano CM, Soares CAS. 2020. First case of SARS-COV-2 sequencing in cerebrospinal fluid of a patient with suspected demyelinating disease. J Neurol 267:3154–3156. 10.1007/s00415-020-09996-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Virhammar J, Kumlien E, Fallmar D, Frithiof R, Jackmann S, Skold MK, Kadir M, Frick J, Lindeberg J, Olivero-Reinius H, Ryttlefors M, Cunningham JL, Wikstrom J, Grabowska A, Bondeson K, Bergquist J, Zetterberg H, Rostami E. 2020. Acute necrotizing encephalopathy with SARS-CoV-2 RNA confirmed in cerebrospinal fluid. Neurology 95:445–449. 10.1212/WNL.0000000000010250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jessen NA, Munk AS, Lundgaard I, Nedergaard M. 2015. The glymphatic system: a beginner’s guide. Neurochem Res 40:2583–2599. 10.1007/s11064-015-1581-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Couderc T, Chretien F, Schilte C, Disson O, Brigitte M, Guivel-Benhassine F, Touret Y, Barau G, Cayet N, Schuffenecker I, Despres P, Arenzana-Seisdedos F, Michault A, Albert ML, Lecuit M. 2008. A mouse model for Chikungunya: young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog 4:e29. 10.1371/journal.ppat.0040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Panciani PP, Saraceno G, Zanin L, Renisi G, Signorini L, Battaglia L, Fontanella MM. 2020. SARS-CoV-2: “three-steps” infection model and CSF diagnostic implication. Brain Behav Immun 87:128–129. 10.1016/j.bbi.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ireland DD, Stohlman SA, Hinton DR, Atkinson R, Bergmann CC. 2008. Type I interferons are essential in controlling neurotropic coronavirus infection irrespective of functional CD8 T cells. J Virol 82:300–310. 10.1128/JVI.01794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dionne KR, Galvin JM, Schittone SA, Clarke P, Tyler KL. 2011. Type I interferon signaling limits reoviral tropism within the brain and prevents lethal systemic infection. J Neurovirol 17:314–326. 10.1007/s13365-011-0038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Detje CN, Meyer T, Schmidt H, Kreuz D, Rose JK, Bechmann I, Prinz M, Kalinke U. 2009. Local type I IFN receptor signaling protects against virus spread within the central nervous system. J Immunol 182:2297–2304. 10.4049/jimmunol.0800596. [DOI] [PubMed] [Google Scholar]
- 127.Griemert EV, Schwarzmaier SM, Hummel R, Golz C, Yang D, Neuhaus W, Burek M, Forster CY, Petkovic I, Trabold R, Plesnila N, Engelhard K, Schafer MK, Thal SC. 2019. Plasminogen activator inhibitor-1 augments damage by impairing fibrinolysis after traumatic brain injury. Ann Neurol 85:667–680. 10.1002/ana.25458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tsai SJ. 2017. Role of tissue-type plasminogen activator and plasminogen activator inhibitor-1 in psychological stress and depression. Oncotarget 8:113258–113268. 10.18632/oncotarget.19935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.St-Jean JR, Jacomy H, Desforges M, Vabret A, Freymuth F, Talbot PJ. 2004. Human respiratory coronavirus OC43: genetic stability and neuroinvasion. J Virol 78:8824–8834. 10.1128/JVI.78.16.8824-8834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hill DP, Robertson KA. 1998. Differentiation of LA-N-5 neuroblastoma cells into cholinergic neurons: methods for differentiation, immunohistochemistry and reporter gene introduction. Brain Res Brain Res Protoc 2:183–190. 10.1016/s1385-299x(97)00041-x. [DOI] [PubMed] [Google Scholar]
- 131.Liu L, Duff K. 2008. A technique for serial collection of cerebrospinal fluid from the cisterna magna in mouse. J Vis Exp 2008(21):960. 10.3791/960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lambert F, Jacomy H, Marceau G, Talbot PJ. 2008. Titration of human coronaviruses using an immunoperoxidase assay. J Vis Exp 2008(14):751. 10.3791/751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kwiatkowska A, Couture F, Levesque C, Ly K, Desjardins R, Beauchemin S, Prahl A, Lammek B, Neugebauer W, Dory YL, Day R. 2014. Design, synthesis, and structure-activity relationship studies of a potent PACE4 inhibitor. J Med Chem 57:98–109. 10.1021/jm401457n. [DOI] [PubMed] [Google Scholar]
- 134.Semaan W, Desbiens L, Houde M, Labonte J, Gagnon H, Yamamoto D, Takai S, Laidlaw T, Bkaily G, Schwertani A, Pejler G, Levesque C, Desjardins R, Day R, D’Orleans-Juste P. 2015. Chymase inhibitor-sensitive synthesis of endothelin-1 (1–31) by recombinant mouse mast cell protease 4 and human chymase. Biochem Pharmacol 94:91–100. 10.1016/j.bcp.2015.02.001. [DOI] [PubMed] [Google Scholar]