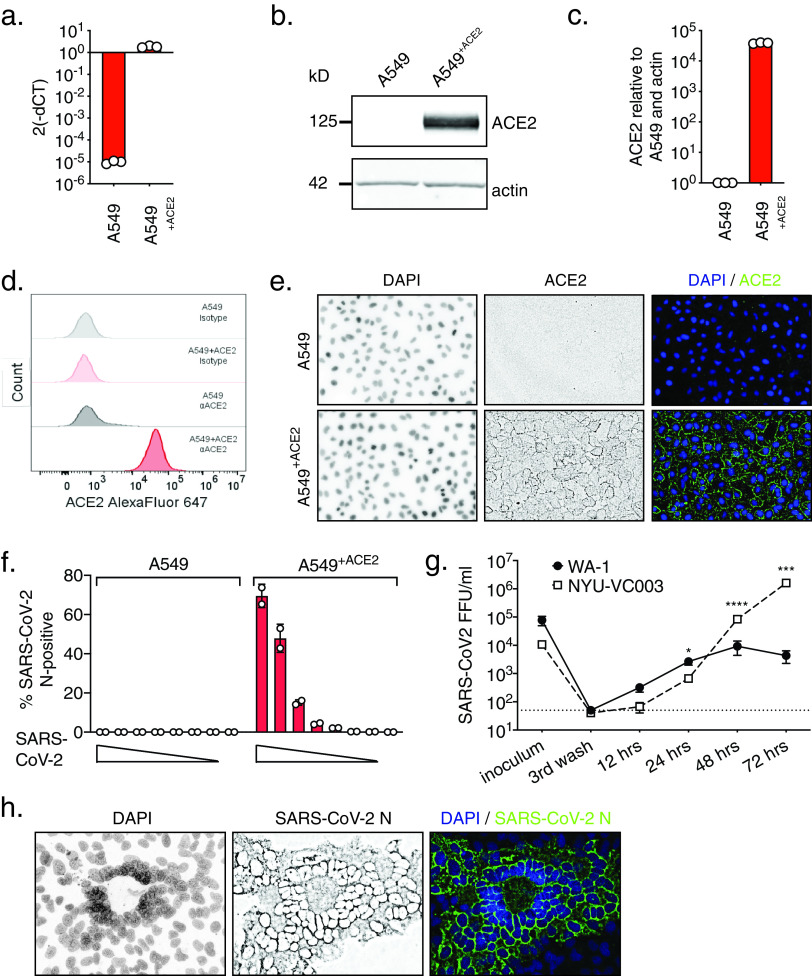
FIG 1.
Validation of A549+ACE2 cells as a tool to study SARS-CoV-2. A549+ACE2 cells were generated by lentiviral transduction delivering an ACE2 overexpression construct and subsequent bulk-selection. (a to e) ACE2 expression in A549 parental or A549+ACE2 cells determined by reverse transcription-quantitative PCR (RT-qPCR) (a); Western blot (b and c), quantified in panel c; flow cytometry (d); or microscopy (e). (f) A549 parental or A549+ACE2 cells were infected with a serial dilution of SARS-CoV-2 USA-WA1/2020. At 24 h, cells were fixed and stained for SARS-CoV-2 N protein, and infected cells were quantified by high-content microscopy. Mean ± standard error of the mean (SEM) from duplicate wells. (g) A549+ACE2 cells were infected with SARS-CoV-2 USA-WA1/2020 or USA/NYU-VC-003/2020 at a multiplicity of infection (MOI) of 0.01, and infectious progeny titers, collected from supernatants over time, determined by focus-forming assay on A549+ACE2 cells. Mean ± SEM from n = 3 independent experiments. Unpaired t test; *, P < 0.05; ***, P < 0.001; ****, P < 0.0001. (h) Confocal microscopy of SARS-CoV-2 syncytia in A549+ACE2 cells at 48 hpi.