HIV-1 persists in individuals on antiretroviral therapy (ART) due to the long-lived cellular reservoirs that contain dormant viruses. Recent discoveries of HIV-1-specific broadly neutralizing antibodies (bNabs) targeting HIV-1 Env protein rekindled the interest in antibody-mediated elimination of latent HIV-1.
KEYWORDS: HIV reservoirs, antibody-dependent killing, human immunodeficiency virus, latency reversal, singly spliced vpu/env mRNA
ABSTRACT
The latent reservoir of HIV-1 is a major barrier for viral eradication. Potent HIV-1 broadly neutralizing antibodies (bNabs) have been used to prevent and treat HIV-1 infections in animal models and clinical trials. Combination of bNabs and latency-reversing agents (LRAs) is considered a promising approach for HIV-1 eradication. PCR-based assays that can rapidly and specifically measure singly spliced HIV-1 vpu/env mRNA are needed to evaluate the induction of the viral envelope production at the transcription level and bNab-mediated reservoir clearance. Here, we reported a PCR-based method to accurately quantify the production of intracellular HIV-1 vpu/env mRNA. With the vpu/env assay, we determined the LRA combinations that could effectively induce vpu/env mRNA production in CD4+ T cells from antiretroviral therapy (ART)-treated individuals. None of the tested LRAs were effective alone. A comparison between the quantitative viral outgrowth assay (Q-VOA) and the vpu/env assay showed that vpu/env mRNA production was closely associated with the reactivation of replication-competent HIV-1, suggesting that vpu/env mRNA was mainly produced by intact viruses. Finally, antibody-mediated killing in HIV-1-infected humanized mice demonstrated that the vpu/env assay could be used to measure the reduction of infected cells in tissues and was more accurate than the commonly used gag-based PCR assay, which measures unspliced viral genomic RNA. In conclusion, the vpu/env assay allows convenient and accurate assessment of HIV-1 latency reversal and bNab-mediated therapeutic strategies.
IMPORTANCE HIV-1 persists in individuals on antiretroviral therapy (ART) due to the long-lived cellular reservoirs that contain dormant viruses. Recent discoveries of HIV-1-specific broadly neutralizing antibodies (bNabs) targeting HIV-1 Env protein rekindled the interest in antibody-mediated elimination of latent HIV-1. Latency-reversing agents (LRAs) together with HIV-1 bNabs is a possible strategy to clear residual viral reservoirs, which makes the evaluation of HIV-1 Env expression upon LRA treatment critical. We developed a PCR-based assay to quantify the production of intracellular HIV-1 vpu/env mRNA. Using patient CD4+ T cells, we found that induction of HIV-1 vpu/env mRNA required a combination of different LRAs. Using in vitro, ex vivo, and humanized mouse models, we showed that the vpu/env assay could be used to measure antibody efficacy in clearing HIV-1 infection. These results suggest that the vpu/env assay can accurately evaluate HIV-1 reactivation and bNab-based therapeutic interventions.
INTRODUCTION
Despite antiretroviral therapy (ART), HIV-1 persists in a small pool of latently infected resting memory CD4+ T cells (1–5). The “shock and kill” approach to purging the latent HIV-1 reservoirs involves pharmacologic reactivation of latent HIV-1 (6). Next, induction of virus-specific host immune responses is required to eliminate infected cells in which HIV-1 gene transcription has been induced by latency reversal agents (LRAs) (7). Antibodies targeting HIV-1 envelope (Env) protein can mediate killing of HIV-1-infected cells through antibody effector functions, such as antibody-dependent cellular cytotoxicity (ADCC) and antibody-dependent cellular phagocytosis (ADCP). In the past 10 years, a variety of bNabs have been isolated from a small subset of HIV-1-positive individuals. bNabs can act against a wide spectrum of viruses by targeting conserved regions on the HIV-1 envelope trimer (8). Recent discoveries of HIV-1-specific bNabs rekindled interest in antibody therapy for HIV-1 prevention as well as for elimination of HIV-1 latent reservoirs. In several phase-I clinical trials, passive administration of a single bNab that included 3BNC117, VRC01, or 10-1074 delayed viral rebound to a level comparable to that in the analytical treatment interruption trials (9–13). bNab infusion accelerated clearance of HIV-1-infected cells via an Fc receptor-dependent mechanism but was not sufficient to clear viral reservoirs (13). Combination therapy with two bNabs that included 3BNC117 and 10-1074 maintained viral suppression after ART discontinuation in patients who did not have preexisting 3BNC117- or 10-1074-resistant viruses (14). Since bNabs cannot target transcriptionally silent HIV-1, LRA and bNab combination for the killing of HIV-1 latently infected cells may be a promising strategy for viral eradication.
The efficacy of LRA and bNab combination therapy is determined by how effectively LRAs could induce HIV-1 Env expression and whether the infused bNabs can target the Env of patient viral isolates. Sensitivity of patient viruses to the chosen bNabs can be measured by in vitro neutralization assays. It is important to quantify HIV-1 Env production induced by LRAs. Although flow cytometry-based measurement of HIV-1 Env expression on the target cell surface is the most accurate method to predict killing efficiency, it is not practical to measure surface HIV-1 Env because of the rarity of latently infected cells and the variability of Env epitopes in patients. PCR-based assays are sensitive and accurate for the measurement of HIV-1 DNA and RNA. Quantitative PCR assays targeting conserved regions of unspliced HIV-1 RNA within gag or pol have been the standard methods to measure LRA-induced cell-associated HIV-1 RNA (15–18). However, the vast majority of HIV-1 proviruses (>95%) in patients under ART are defective, and a significant fraction of the defective viruses carries either entire or truncated gag/pol (19–21). The two major limitations of the gag- or pol-based PCR assay are that (i) it detects some defective viruses that can be transcribed despite carrying lethal mutations or deletions, and (ii) it only detects unspliced viral genomic RNA, which may not be well correlated with the production of spliced viral mRNAs (22). In some studies, spliced HIV-1 tat/rev and env mRNA was measured to evaluate HIV-1 transcription and LRA efficacy (23–25). HIV-1 Vpu and Env are expressed from the same bicistronic mRNA (26). To produce HIV-1 Env protein, induction of HIV-1 multiply spliced tat/rev mRNA and singly spliced HIV-1 vpu/env mRNA are required. Expression of HIV-1 Env protein requires leaky scanning of bicistronic vpu/env mRNA, thereby allowing ribosomes access to the downstream env open reading frame (27). Another advantage for detecting vpu/env transcripts is that they are more likely produced from intact proviruses, but not from defective proviruses, because more than 90% of the defective HIV-1 proviruses in patients carry deletions or splice site mutations in the vpu/env open reading frame (ORF) (20, 21, 28). Here, we report a quantitative PCR-based assay to specifically measure singly spliced HIV-1 vpu/env mRNA. This vpu/env assay can be used to evaluate the production of HIV-1 Env at the transcriptional level and evaluate bNab-mediated killing of infected cells in which HIV-1 latency is reversed by LRAs.
RESULTS
Measurement of singly spliced HIV-1 vpu/env transcripts.
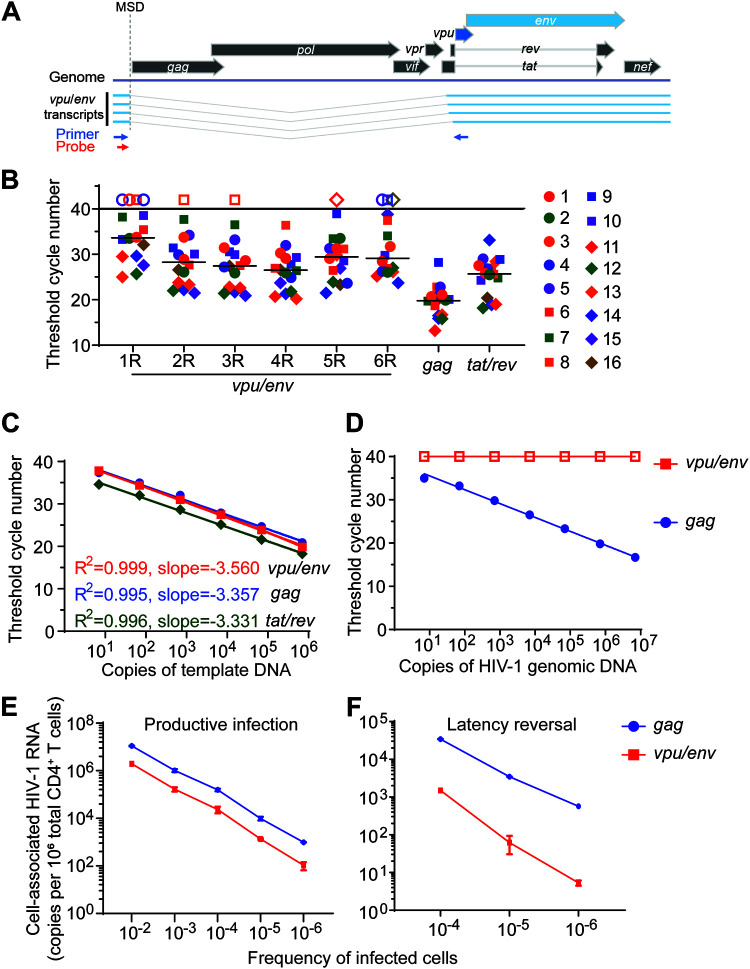
HIV-1 Vpu and Env are expressed from the same bicistronic mRNA, and HIV-1 vpu/env mRNA utilizes the major splicing donor site (MSD) and A4 or A5 splicing acceptor site (29). To design primers and probes for the detection of HIV-1 vpu/env mRNA, the 5′ primer should be located upstream of the MSD, while the 3′ primer should be downstream of the A5 splicing acceptor site to account for all the vpu/env splicing variants (Fig. 1A). The length between 5′ primer and the env start codon is about 310 to 350 bp, which is much larger than the ideal size of a quantitative PCR (qPCR) amplicon (70 to 200 bp); qPCR efficiency would be low if the 3′ primer is in the env ORF. Therefore, we placed the 3′ primer within conserved regions of the vpu ORF, based on the HIV Sequence Compendium 2018 (www.hiv.lanl.gov), and designed six primer/probe sets to measure vpu/env mRNA (Table 1). Ten clade B HIV-1 isolates (NIH AIDS Reagent Program) and six viral isolates from HIV-1-positive individuals were used for assay optimization. HIV-1 gag- or tat/rev-based PCRs were used as controls. qPCR results showed that vpu/env mRNA was detected in all samples using the no. 4 primer set (Fig. 1B). HIV-1 gag and tat/rev mRNA were also detected in all samples (Fig. 1B). Therefore, the no. 4 primer set was selected for the following studies. Next, standard plasmids containing the vpu/env, gag, or tat/rev PCR fragments were measured separately with corresponding primer/probe sets to confirm the assay sensitivity (Fig. 1C). The 95% confidence intervals for the slope of each assay are −3.637 to −3.482 (vpu/env), −3.526 to −3.188 (gag), and −3.487 to −3.176 (tat/rev). The vpu/env assay only targets singly spliced vpu/env RNA transcripts, but not unspliced viral genomic RNA or proviral DNA. As predicted, no signal could be detected by the vpu/env assay, even with the presence of 1 × 107 copies of HIV-1 genomic DNA (Fig. 1D). Together, these results demonstrated that the vpu/env assay had great sensitivity and specificity for the spliced vpu/env RNA, but not for viral genomic RNA or proviral DNA.
FIG 1.
Sensitivity and specificity of the vpu/env assay. (A) The singly spliced HIV-1 vpu/env mRNA. Position of primers (blue arrow) and probe (red arrow) of the vpu/env assay. (B) Evaluation and comparison of vpu/env-specific primers. CD4+ T cells were infected by 10 HIV-1 clade B isolates (1–10) and viruses isolated from six HIV-1-positive patients (11–16). Total RNA was used for vpu/env mRNA detection. Each of the six 3′ primers was paired with the same 5′ primer and probe. Threshold cycle (CT) numbers are shown. Open symbols and horizontal lines represent no detection and the median CT values, respectively. (C) Assay sensitivity. Serially diluted vpu/env, gag, and tat/rev standard DNA were measured separately in duplicate by quantitative PCR (qPCR). (D) Assay specificity. Serially diluted pNL4-3 was used as the template for qPCR. Open squares, undetected. (E and F) Detection of intracellular HIV-1 mRNA. Intracellular HIV-1 mRNA in serially diluted infected cells. Productively (E) or latently (F) infected primary CD4+ T cells were serially diluted in uninfected CD4+ T cells. For latent infection, cells were then treated with phorbol myristate acetate (PMA)-ionomycin for 24 h. HIV-1 vpu/env and gag mRNA were measured by reverse transcription-quantitative PCR (RT-qPCR).
TABLE 1.
List of probes and primers for vpu/env RT-qPCR
| Probe or primer purpose | Probe or primer type | Sequence (5′–3′)a |
|---|---|---|
| gag qPCR | Probe | VIC-CTATCCCATTCTGCAGCTTCCTCATTGATG-TAMRA |
| Forward primer | ACATCAAGCAGCCATGCAAAT | |
| Reverse primer | TCTGGCCT GGTGCAATAGG | |
| tat/rev qPCR | Probe | VIC-TTCCTTCGGGCCTGTCGGGTCCC-TAMRA |
| Forward primer | CTTAGGCATCTCCTATGGCAGGAA | |
| Reverse primer | GGATCTGTCTCTGTCTCTCTCTCCACC | |
| vpu/env qPCR | Probe | FAM- CGCACRGCAAGAGGCGAGGG-MGB |
| Forward primer | AGCTCTCTCGACGCAGGACTC | |
| R1 reverse primer | TACTACTYACTGCTTTGRTAGA | |
| R2 reverse primer | CATYACATGTACTACTYACTGCTTTG | |
| R3 reverse primer | GCATYACATGTACTACTYACTGCTTT | |
| R4 reverse primer | AAAGGTTGCATTACATGTACTACTTACTGCTTT | |
| R5 reverse primer | GCTACTACTAATGCTACTATTGCTAATAT | |
| R6 reverse primer | GCTAATATTTGTAAAGGTTGCATTACATGTACTAC |
TAMRA, 6-carboxytetramethylrhodamine; FAM, 6-carboxyfluorescein. The underlined probe and primer were chosen for the vpu/env assay.
Next, we sought to determine whether this assay could detect vpu/env mRNA in samples with low frequencies of HIV-1-infected cells. CD4+ T cells infected with the HIV-1 reporter virus NL4-3-ΔEnv-EGFP were serially diluted with uninfected cells. In samples containing 1 HIV-1-infected cell out of 1 million total cells, about 100 copies of intracellular vpu/env mRNA were detected, which was 10-fold lower than the unspliced viral genomic RNA detected by the gag assay (Fig. 1E). Reactivation of latent HIV-1 by LRA is the first step of the “shock and kill” strategy (30–32). To test if the vpu/env assay can detect reactivated viruses, we generated latently infected cells in vitro using a previous described reporter virus (33). Cells containing known percentages of latent HIV-1 were serially diluted into uninfected CD4+ T cells to achieve the low frequency of latently infected cells. After latency reversal by phorbol myristate acetate (PMA)-ionomycin, vpu/env mRNA could be detected in samples containing as little as a single latent HIV-1 in 1 million cells (Fig. 1F). Taken together, these experiments demonstrate that the vpu/env assay is highly specific and sensitive and is capable of capturing low levels of vpu/env mRNA production after viral latency reversal.
Abundance of vpu/env mRNA correlates with expression of Env protein.
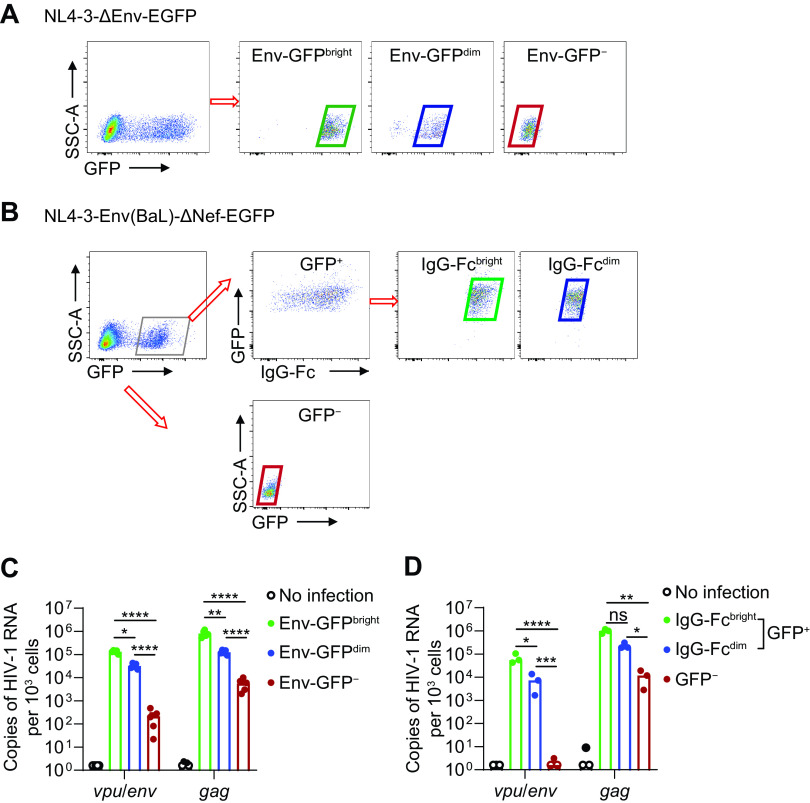
Next, we determined whether the vpu/env assay measuring vpu/env transcripts can be used to evaluate the production of HIV-1 Env protein, especially on the surface of infected cells. CD4+ T cells were infected with NL4-3-ΔEnv-EGFP in which env was disrupted by inserted egfp. Thus, enhanced green fluorescent protein (EGFP) expression mirrored total Env protein level. We found that both vpu/env and gag mRNA correlated well with EGFP expression and were detectable even in cells without EGFP expression (Fig. 2A and C), suggesting some discordance between viral gene transcription and EGFP expression. To further determine the correlation between vpu/env mRNA level and surface expression of Env protein, we infected CD4+ T cells with NL4-3-Env(Ba-L)-Δnef-EGFP, which encoded Env of HIV-1Ba-L and contained an EGFP coding gene inserted in nef. We used bNab PGT121 and anti-human IgG-Fc antibodies sequentially to detect cell surface Env protein. Green fluorescent protein-positive (GFP+) cells were purified to exclude uninfected cells coated with free GP120 because of its shedding from infected cells. The abundance of vpu/env but not gag transcripts correlated very well with cell surface Env expression (Fig. 2B and D). These results demonstrated that the vpu/env assay can be used to quantitatively measure the change of cell surface HIV-1 Env protein.
FIG 2.
Correlation between HIV-1 vpu/env mRNA level and Env protein level. (A) Sorting strategy for Env-GFPBright, Env-GFPdim, and Env-GFP− cells. Activated CD4+ T cells were infected with NL4-3ΔEnv-EGFP. Env-GFPBright, Env-GFPdim, and Env-GFP− cells were sorted based on green fluorescent protein (GFP) intensity on day 3 postinfection. (B) Sorting strategy for IgG-FcBright, IgG-Fcdim, and GFP− cells. Activated CD4+ T cells were infected with NL4-3-Env(Ba-L)-Δnef-EGFP. On day 3 postinfection, surface Env protein was labeled by PGT121 followed with anti-human IgG Fc antibodies. (C and D) Cell-associated vpu/env or gag mRNA was measured by qPCR. P values were calculated by one-way analysis of variance (ANOVA) with Tukey’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Detection of vpu/env mRNA in LRA-treated CD4+ T cells from patients on ART.
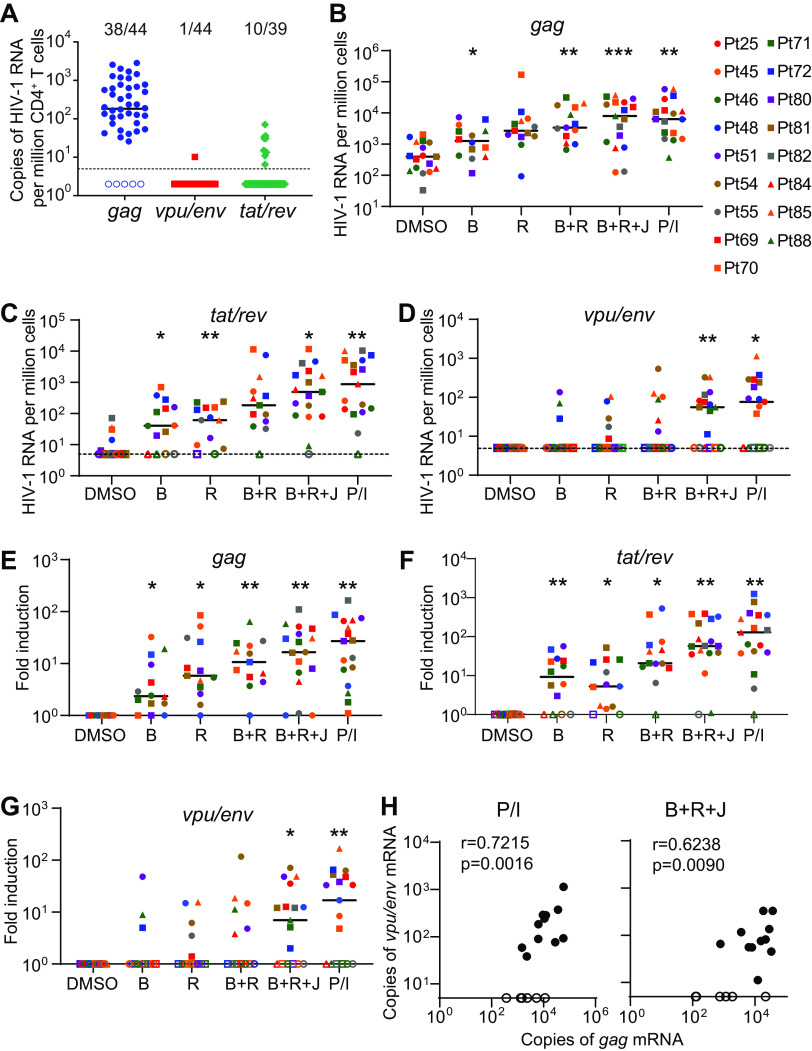
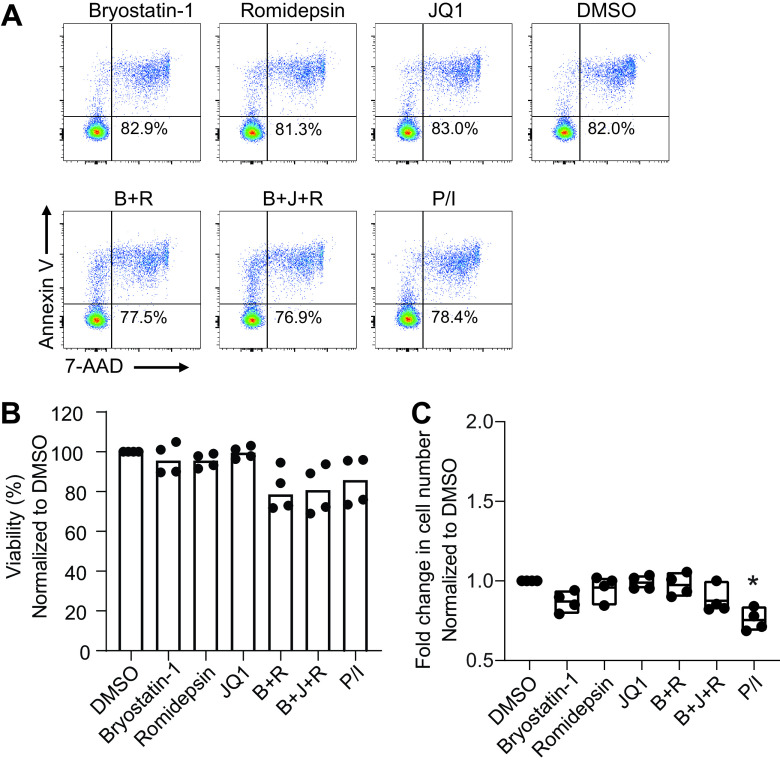
The gag assay that detects unspliced HIV-1 genomic RNA is the most commonly used method to evaluate HIV-1 latency reversal (15, 34). We performed gag, vpu/env, and tat/rev assays to compare the levels of unspliced, singly spliced, and multiply spliced HIV-1 transcripts in unstimulated CD4+ T cells from 44 patients on ART. The unspliced viral genomic RNA (gag) was detectable in 38/44 samples, much more frequently than vpu/env (1/44) and tat/rev mRNA (10/39) (Fig. 3A). Unspliced HIV-1 genomic RNA can be produced by defective but transcriptionally active HIV-1 that does not trigger cell death or immune recognition. It may also be derived from chimeric host-HIV-1 transcripts because the virus integrates into actively transcribed host genes (35, 36). To evaluate the induction of vpu/env mRNA, patient CD4+ T cells were treated with various LRAs for less than 24 h. Bryostatin-1 or romidepsin alone can induce unspliced HIV-1 RNA (18, 37). In our study, induction of gag mRNA (fold change, >1) by single LRAs, as well as by LRA combinations, was observed in most individuals (Fig. 3B and E). The magnitude of increase in tat/rev mRNA was comparable to that of gag mRNA in different LRA groups (Fig. 3C and F). In contrast, significant induction of vpu/env mRNA was only achieved when (i) a combination of bryostatin-1, JQ1, and romidepsin or (ii) PMA-ionomycin was used (Fig. 3D and G). The induction of gag and vpu/env mRNA was positively correlated in the PMA-ionomycin and the triple LRA combination groups (Fig. 3H). In the LRA experiments, patient CD4+ T cells were treated for less than 24 h, and no significant cytotoxicity was observed in single LRA groups (Fig. 4), which excluded the possibility that the lack of detection of HIV-1 transcripts by single LRAs was due to cytotoxicity. Single LRAs had poor induction of vpu/env mRNA in most samples, indicating the presence of multiple blocks in HIV-1 transcription in latently infected cells, such as sequestration of essential transcription factors like nuclear factor of activated T cells (NFAT) and NF-κB in the cytoplasm, transcriptional interference by the upstream portion of the host gene, DNA methylation, histone deacetylation, and restrictive chromatin structures (38–41).
FIG 3.
Quantification of HIV-1 vpu/env mRNA in blood CD4+ T cells from patients under suppressive antiretroviral therapy (ART). (A) Spliced and unspliced cell-associated HIV-1 RNA in freshly isolated CD4+ T cells without latency reversal from 44 patients. HIV-1 gag, vpu/env, and tat/rev mRNA was measured by RT-qPCR using total RNA from 0.5 million patient CD4+ T cells. (B to G) Reactivation of latent HIV-1 by latency-reversing agents (LRAs) or LRA combinations. Patient CD4+ T cells were treated with different LRAs or combination for 18 to 24 h. Virus reactivation was determined by quantitative measurement of indicated HIV-1 transcripts. Total RNA from 0.5 million patient CD4+ T cells was used for the gag, vpu/env, or tat/rev qPCR assays. Copies of viral transcripts (B to D) and fold induction (E to G) in each treatment group are shown. P values were calculated by one-way ANOVA with Tukey’s test. *, P < 0.05; **, P < 0.01. (H) Correlation between copies of gag and vpu/env transcripts after latency reversal. Values in panels B and D were used. r and P values were determined by Spearman correlation. DMSO, dimethyl sulfoxide; B, bryostatin-1; J, JQ1; R, romidepsin; P/I, PMA plus ionomycin; open symbols, undetectable.
FIG 4.
Toxicity of LRA to CD4+ T cells. (A and B) Viability of CD4+ T cells after LRA treatment. Unstimulated CD4+ T cells from four heathy donors were treated with single LRAs or LRA combination (3 million cell/group) for 18 h. Cell viability was measured by annexin V and 7-aminoactinomycin D (7-AAD). (C) Cell counts in each treatment group. P values were calculated by one-way ANOVA. *, P < 0.05.
Induction of vpu/env mRNA is correlated with reactivation of replication-competent latent HIV-1.
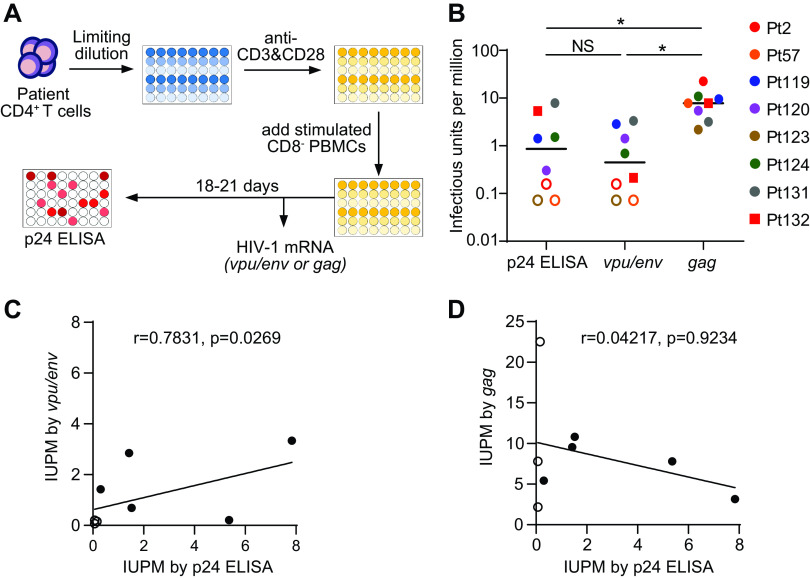
PCR-based measurement of HIV-1 transcripts relies on detecting a small region of the viral genome and does not distinguish intact and defective proviruses. Interestingly, a recent study demonstrated that the majority of the defective HIV-1 proviruses carried mutations or deletions that abolished the production of singly spliced vpu/env transcripts (21). It is possible that the vpu/env assay preferentially detects viral transcripts from replication-competent viruses. To evaluate if this assay can measure reactivation of replication-competent viruses, we determined the frequency of latent HIV-1 after virus reactivation by the standard quantitative viral outgrowth assay (Q-VOA) and the vpu/env assay (Fig. 5A). The infectious units per million CD4+ T cells (IUPM) determined by the vpu/env assay (median, 0.45; range, 0.07 to 3.33) were positively correlated with the values determined by the Q-VOA (median, 0.86; range, 0.07 to 7.84) (Fig. 5B and C). In contrast, the IUPM values determined by gag-based qPCR were much higher (median IUPM, 7.80; range, 2.18 to 22.54) (Fig. 5B and D), primarily due to viral transcripts produced by replication-defective viruses. Our results demonstrated that the spliced vpu/env mRNA is a better surrogate than the full-length viral genomic RNA to evaluate the reactivation of replication-competent latent HIV-1. Since we could not exclude the possibility that the vpu/env assay detected transcripts from defective proviruses, it is helpful to monitor the exponential increase in vpu/env transcripts from the same wells over time. Nonetheless, the vpu/env assay can accurately predict IUPM values on day 7, making it a rapid assay to determine the size of HIV-1 reservoirs.
FIG 5.
Production of vpu/env mRNA is correlated with reactivation of replication-competent HIV-1. (A) Scheme of quantitative viral outgrowth assay. At day 0, CD4+ T cells were isolated from patient blood. Cells were seeded by serial dilutions and stimulated with anti-CD3/CD28 for 3 days. After costimulation, PHA-stimulated CD8− PBMCs from healthy donors were added to support viral outgrowth. At day 7, half of the cells were harvested for HIV-1 mRNA measurement. The remaining cells were cultured for another 11 to 14 days before p24 enzyme-linked immunosorbent assay (ELISA). (B) Infectious units per million CD4+ T cells (IUPM) measurement by different assays. HIV-1 infection was determined by p24 ELISA and the vpu/env or gag assay. IUPM was calculated by the frequency of p24+ or HIV-1 mRNA+ wells, as previously described (44). P values were calculated by one-way ANOVA with Tukey’s test. *, P < 0.05. (C and D) Correlation of p24 ELISA and the PCR-based assays. r and P values were determined by Spearman correlation. Open symbols indicate undetectable.
Antibody-mediated killing of infected cell can be measured by the vpu/env assay.
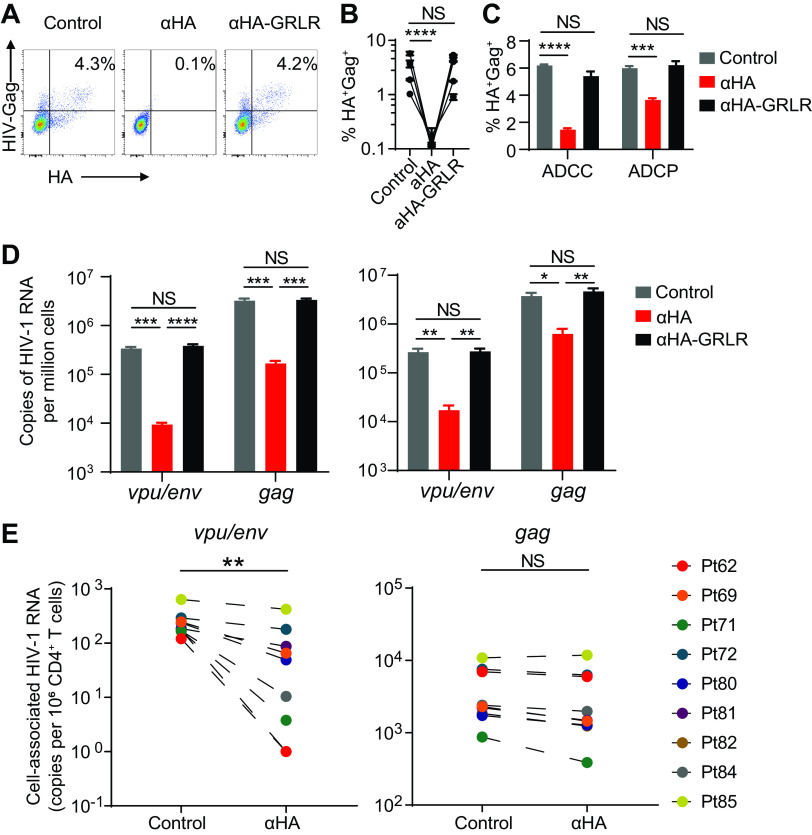
Antibodies suppress HIV-1 infection by neutralization of cell-free viruses and killing of virus-infected cells through ADCC and ADCP (42). Antibody-mediated killing of infected cells relies on the expression of HIV-1 Env on the target cells, which are vpu/env mRNA+ cells. We hypothesize that vpu/env mRNA measurement could be used to evaluate antibody efficacy against persistent HIV-1 reservoirs. To test this hypothesis, we used a recombinant HIV-1 reporter virus (HIVivoHA) expressing a murine heat-stable antigen (HSA) bearing a hemagglutinin (HA) tag (43). Anti-HA antibodies can trigger antibody-dependent killing by binding to the HA on the surface of infected cells. We infected CD8-depleted healthy donor peripheral blood mononuclear cells (PBMCs) with the reporter virus in the presence or absence of anti-HA antibodies. Anti-HA antibodies cleared HIVivoHA- infected cells (Fig. 6A and B). To confirm that the cell killing was Fc-dependent, a modified anti-HA antibody with mutations that abrogate Fc receptor binding (anti-HA:GRLR) was generated, which failed to clear infected cells (Fig. 6A and B). In this culture system, we used CD8-depleted PBMCs that contained both NK cells and monocytes. We further confirmed that both NK cells and monocytes could mediate Fc-dependent cell killing through ADCC and ADCP, respectively (Fig. 6C).
FIG 6.
Antibody-mediated killing of infected cells can be accurately measured by vpu/env RT-qPCR. (A and B) Antibody-mediated clearance of HIV-1-infected cells. Healthy donor PBMCs were infected with the HIVivoHA reporter virus. After antibody treatment, residual infected cells (HA+ Gag+ cells) were determined by flow cytometry at day 4 postinfection. Experiments were performed using PBMCs from four different donors. (C) Antibody-dependent cellular cytotoxicity (ADCC) and antibody-dependent cellular phagocytosis (ADCP). HIVivoHA-infected CD4+ T cells were cocultured with purified autologous NK cells and monocyte-derived-macrophages at a 1:1 ratio for 6 h with the presence of indicated antibodies. Remaining live infected cells were determined by flow cytometry. (D and E) Antibody-mediated clearance of HIV-1 mRNA+ cells. (D) Experiments were performed using PBMCs from two healthy donors. After antibody treatment, residual infected cells were used to measure cell-associated vpu/env or gag RNA. (E) The same antibody treatment experiments were performed as described in panels A to D. After anti-hemagglutinin (HA) treatment, remaining in vitro-infected cells were diluted with blood CD4+ T cells from 9 HIV-1-positive patients right before cellular RNA extraction. vpu/env mRNA and gag mRNA were measured by qPCR. In panels B, C, and D, error bars show mean values with standard error of the mean (SEM). P values were calculated by one-way ANOVA with Tukey’s test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. In panel E, P values were calculated by paired t test. **, P < 0.01.
Having confirmed clearance of infected cells by flow cytometry, we performed the same in vitro antibody killing assay shown in Fig. 6A and measured intracellular viral mRNA by the vpu/env and gag assays. We observed significant decreases of HIV-1 mRNA upon anti-HA antibody treatment by both vpu/env and gag assays (Fig. 6D). While defective viruses do not accumulate during short-term in vitro infection, HIV-1 proviruses in patient samples are predominantly defective and produce mutated or truncated viral transcripts upon latency reversal. Antibodies cannot clear the vast majority of HIV-1 RNA+ cells in patients who do not have an intact vpu/env ORF. Therefore, it is important to assess whether the vpu/env assay could accurately quantify the clearance of vpu/env mRNA+ cells by antibodies without being affected by the overwhelming presence of the defective viruses. When antibody-treated or control HIVivoHA-infected cells were spiked into patient CD4+ T cells (<5 infected cells in 1 million patient cells) to mimic the frequency of reactivable intact HIV-1 in patients, the gag assay was not able to sense the antibody effect, because the mutant viral transcripts from patient CD4+ T cells vastly outnumbered the ones produced by HIVivoHA-infected cells. In contrast, the reduction of infected cells by antibodies was detectable by the vpu/env assay (Fig. 6E). Our results suggest that the vpu/env assay is more sensitive and accurate to denote the reduction of HIV-1-infected cells from patients.
Assessment of HIV-1 bNab efficacy by the vpu/env assay in humanized mice.
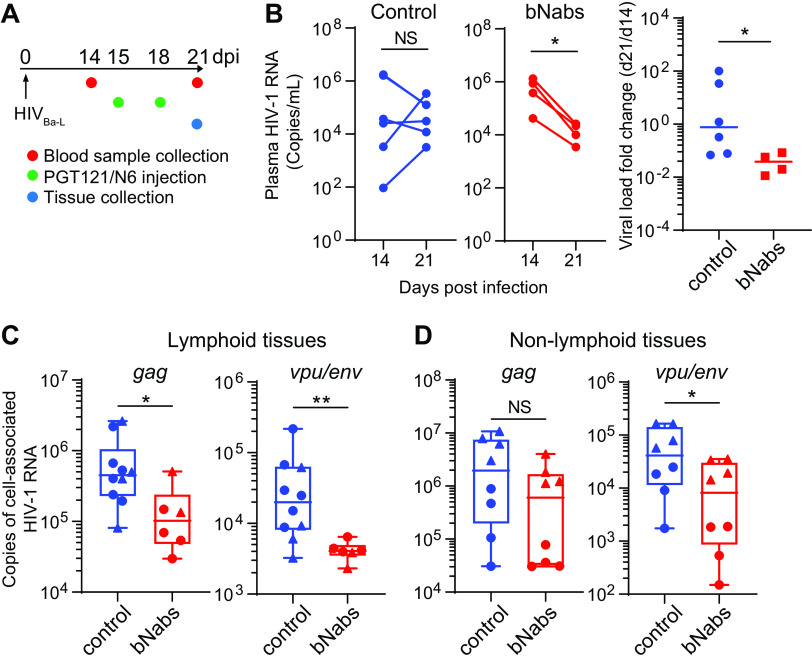
The gag assay is commonly used to quantify cell-free plasma HIV-1 levels and cell-associated viral RNA in patients and experimental animals. However, defective HIV-1 rapidly accumulates during acute infection in patients (20, 21) and humanized mice (44, 45). Passive administration of a single HIV-1-specific bNab in humanized mice resulted in a small and transient reduction in viremia (45). Treatment with bNab combinations led to prolonged viral suppression in viremic mice and delayed viral rebound from aviremic mice (45, 46). To evaluate whether the vpu/env assay could be more sensitive and accurate in quantifying HIV-1 in experimental animals under bNab therapy, we infected humanized MISTRG-6-15 mice with HIV-1Ba-L and then treated the mice with a combination of two HIV-1 bNabs, PGT121 and N6 (Fig. 7A). Antibody therapy started at day 15 postinfection, when plasma viral load peaked. The levels of plasma HIV-1 RNA had an average decrease of 23.3-fold in bNab-treated mice (Fig. 7B). To determine bNab efficacy in tissues, cell-associated viral RNA was quantified by the vpu/env and gag assays. A significant reduction of vpu/env mRNA was observed in lymphoid tissues (spleen and lymph node) and nonlymphoid tissues (liver and lung), while gag mRNA was modestly reduced in some tissues (Fig. 7C and D). This discrepancy is probably attributed to the accumulation of defective HIV-1 proviruses in tissues, which could not be cleared by bNabs. The results from bNab-treated humanized mice suggest that the vpu/env assay can more accurately quantify cell-associated HIV-1 RNA and evaluate antibody efficacy in vivo, when specimens contain defective HIV-1.
FIG 7.
Quantification of vpu/env mRNA accurately assesses bNab efficacy in HIV-1-infected humanized mice. (A) Treatment scheme. (B and C) bNab treatment suppresses HIV-1 replication. MISTRG-6-15 mice engrafted with human cord blood CD34+ cells were infected with HIVBa-L. Plasma viral load was measured at days 14 and 21 postinfection. (C and D) Quantification of cell-associated gag and vpu/env RNA. In panel C, cell-associated HIV-1 RNA was measured in spleens (circle) or lymph nodes (triangle) of control or bNab-treated mice. In panel D, lungs (triangle) or livers (circle) were collected for the measurement of cell-associated HIV-1 RNA. (B) P values were calculated by paired t test. (C, D, and E) P values were calculated by the Mann-Whitney U test. *, P < 0.05; **, P < 0.01.
DISCUSSION
The combination of LRAs and bNabs is considered a promising approach to reduce or eliminate latent HIV-1 in patients on suppressive ART (31, 47). To evaluate the efficacy of latency reversal and bNab-mediated killing, a quantitative method for HIV-1 Env expression is needed. HIV-1 mRNA is often measured by reverse transcription-quantitative PCR (RT-qPCR) using gag or pol-specific primers (15–18). However, these methods only detect unspliced HIV-1 mRNA. The unspliced viral mRNA in patient CD4+ T cells is detectable by the gag assay without LRA treatment. It is likely from three difference sources, namely chimeric HIV-1/host transcripts, defective but transcriptionally active viruses, or sporadic reactivation of latent HIV-1. The vast majority of HIV-1 proviruses (>95%) in patients under ART are defective and are not able to cause viral rebound (19–21). The vpu/env assay can disregard the vast majority of the transcripts from defective proviruses, because the majority of the HIV-1 proviruses in patients carry deletions or splice site mutations in vpu and env (15–18). We acknowledge that the vpu/env assay can detect defective viruses which have intact Vpu and functional MSD and A4 or A5 splicing acceptor sites. Such defective viruses are probably rare and do not affect the quantification of replication-competent viruses by the vpu/env assay (Fig. 5). It is important to compare the vpu/env assay with other latent HIV-1 quantification methods, including an alternative version of Q-VOA using RT-PCR as the readout (48), tat/rev-induced limiting dilution assay (TILDA) (24), and intact proviral DNA assay (IPDA) (21). Although the vpu/env assay cannot replace the gag assay to measure cell-free HIV-1, it is a better assay to quantify cell-associated HIV-1 RNA for in vivo studies due to the presence of defective viruses (Fig. 6 and 7). In conclusion, we developed and validated a PCR-based assay to measure HIV-1 vpu/env mRNA. The vpu/env assay is sensitive and highly specific for HIV-1 vpu/env mRNA. It can serve as an alternative to existing assays to quantify cell-associated HIV-1 RNA. This assay is better suited for the evaluation of LRAs and bNab-based HIV-1 cure strategies.
MATERIALS AND METHODS
Plasmids, antibodies, and viruses.
The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: N6 heavy chain- and light chain-expressing plasmids (no. 12967 and 12966, from J. Huang and M. Connors), pNL4-3 (no. 114, from M. Martin), pNL4-3ΔEnv-enhanced green fluorescent protein (EGFP) (no. 11100, from H. Zhang, Y. Zhou, and R. Siliciano), HIV-1Ba-L (no. 510, from S. Gartner, M. Popovic, and R. Gallo), and 10 HIV-1 clade B isolates (no. 11412). HA antibody heavy chain- and light chain- expressing plasmids, as well as pHIVivoHA, were kindly provided by Michel Nussenzweig. Codon-optimized PGT121 heavy chain- and light chain-expressing plasmids were kindly provided by Dennis Burton. PGT12, N6, and anti-HA antibodies were produced by transfecting the FreeStyle 293-F cells (Thermo Fisher). pNL4-3-Δ6-drEGFP and the packaging vector pC-Help were kindly provided by Robert Siliciano. HIVivoHA and NL4-3-Env(Ba-L)-Δnef-EGFP were prepared by transfecting HEK293T with pHIVivoHA and pNL4-3-Env(Ba-L)-Δnef-EGFP. NL4-3ΔEnv-EGFP was prepared with viral vector and NL4-3 envelope-expressing plasmid. NL4-3-Δ6-drEGFP was prepared with viral vector, NL4-3 envelope-expressing plasmid, and pC-Help. For animal infection, HIV-1Ba-L was prepared from phytohemagglutinin (PHA)-stimulated CD8-depeleted PBMCs from healthy donors. Concentrated viral stocks were prepared using the Lenti-X concentrator (TaKaRa).
Flow cytometry analysis.
Antibodies, which included mouse CD45 (clone 30-F11), human CD45 (clone HI30), human CD3 (clone HIT3a), human CD4 (clone OKT4), human CD8 (clones HIT8a and RPA-T8), human CD14 (clone M5E2), human NKp46 (clone 9E2), and human IgG Fc (clone QA19A42), were purchased from BioLegend. Anti-HA antibody (clone GG8-1F3.3.1) was purchased from Miltenyi Biotec. Anti-HIV-1 p24 (lone KC57-RD1) was purchased from Beckman Coulter, and anti-HIV-1 p24 (clone KC57-FITC) was obtained from the NIH AIDS Reagent Program. For intracellular staining, cells were fixed with Cytofix/Cytoperm (BD Biosciences). Annexin V and 7-aminoactinomycin D (7-AAD) viability staining solution (BioLegend) were used for the detection of dead and apoptotic cells.
Human subjects.
HIV-1-positive adult patients were recruited by the Infectious Diseases Clinic at Barnes-Jewish Hospital. Both male and female patients were included. All patients were on ART and had maintained undetectable plasma HIV-1 RNA levels (<20 copies per ml) for at least 6 months prior to blood collection. This study was approved by Washington University School of Medicine Internal Review Board. All study participants were provided with written informed consent. To obtain PBMCs for in vitro studies, anonymous peripheral blood samples were acquired from the Mississippi Valley Regional Blood Center as waste cellular products. Human PBMCs or purified CD4+ T cells were cultured in RPMI 1640 medium containing 10% fetal bovine serum at 37°C and 5% CO2. CD4+ T cells were isolated using the EasySep human CD4+ T-cell isolation kit (Stemcell Technologies).
In vitro HIV-1 infection.
CD4+ T cells from healthy donors were stimulated with anti-CD3 antibody at 1 μg/ml−1 (BioLegend), anti-CD28 antibody at 1 μg/ml (BioLegend), and 20 ng/ml IL-2 (BioLegend) for 3 days. Activated CD4+ T cells were infected with 10 clade B HIV-1 isolates and six clinical isolates, with NL4-3ΔEnv-EGFP. Then cells were cultured with 20 ng/ml interleukin 2 (IL-2) for 2 to 3 days. To generate latently infected cells, activated CD4+ T cells were infected with NL4-3-Δ6-drEGFP and cultured with 2 ng/ml IL-2 for 2 weeks. Then, GFP-negative (GFP−) cells were sorted and reactivated by 50 ng/ml PMA (Sigma) plus 1 μM ionomycin (Sigma) for 24 h.
Latency reversal agents and in vitro treatment.
Patient blood CD4+ T cells were treated in the presence of 10 μM T-20 (AIDS Reagent Program no. 9409) and 5 μM raltegravir (AIDS Reagent Program no. 11680) with latency-reversing agents (LRAs) at the following concentrations: 10 nM bryostatin-1 (Sigma), 1 μM JQ1 (Selleck), 40 nM romidepsin (Sigma), and 50 ng/ml PMA (Sigma) plus 1 μM ionomycin (Sigma), or with medium alone plus dimethyl sulfoxide (DMSO) for all single and combination treatments. The concentrations of LRAs were chosen based on a previously published in vitro study (18). No fewer than 2.5 million cells were used in each group of cells. Because of variable yields of CD4+ T cells, a few agents were not tested in all individuals. After 18 to 24 h of LRA treatment, cells of each groups were harvested and lysed with 0.6 ml of TRIzol reagent (Invitrogen) for the quantification of HIV-1 RNA transcripts.
Measurement of cell-associated HIV-1 RNA.
To quantify cell-associated HIV-1 RNA, total cellular RNA was isolated using the Direct-zol RNA Miniprep Plus kit (Zymo Research), which incorporates a DNase I digestion step to eliminate cellular DNA. First-strand cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s procedures. Real-time PCR was performed using TaqMan Universal PCR master mix (Applied Biosystems) on an ABI QuantStudio3 real-time PCR machine. The qPCR conditions are listed in Table 2. To quantify HIV-1 RNA in infected cells in vitro, each qPCR mixture contained cDNA generated from 0.5 million total CD4+ T cells. To measure latency reversal in patient samples, each PCR mixture contained cDNA generated from 0.5 million patient CD4+ T cells. The detection limit for the qPCR assays is 5 copies of HIV-1 RNA per million cells. Primers and probes used for HIV-1 gag and tat/rev mRNA measurement were described previously (15, 23). Probes and primers are listed in Table 1. Reverse primer 4R was chosen in this study.
TABLE 2.
qPCR conditions
| qPCR component or condition | Concn |
Cycle length | No. of cycles | |
|---|---|---|---|---|
| Stock | Final | |||
| TaqMan Universal PCR master mix | 2× | 1× | ||
| Forward primer | 10 μM | 500 nM | ||
| Reverse primer | 10 μM | 500 nM | ||
| Probe | 2.5 μM | 125 nM | ||
| Cycle temperature | ||||
| 95°C | 2 min | |||
| 95°C | 2 s | 40 | ||
| 60°C | 20 s | 40 | ||
Generation of qPCR standard.
To generate a DNA standard for qPCR assays, primary CD4+ T cells were infected with HIVNL4-3. Total cellular RNA was extracted for reverse transcription. The cDNA fragments amplified by gag-, vpu/env-, or tat/rev-specific PCR primers were introduced into a TOPO-TA cloning plasmid (Thermo Fisher). The TOPO-TA cloning plasmids containing the PCR fragment were used as a qPCR standard.
Quantitative viral outgrowth assay.
Q-VOA was adapted as previously described (49). On day 0, CD4+ T cells from HIV-1-positive individuals on suppressive ART were isolated and plated at 1.0 × 106, 0.5 × 106, and 0.2 × 106 cells/well before stimulation with anti-CD3/CD28 antibodies for 3 days. CD8-depleted PHA-stimulated PBMCs from healthy donors were added at day 0 and day 7 after costimulation. Briefly, CD8+ T cells were depleted using MojoSort human CD8 nanobeads (BioLegend) and subsequently stimulated with 20 ng/ml IL-2 (BioLegend) and 17 μg/ml PHA-L (Sigma) for 72 h. At day 7, half of the cells were harvested for quantification of HIV-1 gag and vpu/env mRNA. The remaining cells were cultured for another 11 to 14 days before collecting culture supernatant for HIV-1 p24 ELISA (XpressBio).
Correlation analysis of vpu/env transcripts and Env expression in HIV-1-infected cells.
Activated primary CD4+ T cells were infected with NL4-3ΔEnv-EGFP and cultured with 20 ng/ml IL-2 for 3 days. Env-GFPBright, Env-GFPdim, and Env-GFP− cells were sorted based on GFP intensity. To isolate cells with surface Env expression, activated primary CD4+ T cells were infected with NL4-3-Env(Ba-L)-Δnef-EGFP and cultured with 20 ng/ml IL-2 for 3 days. Infected cells were incubated with PGT121 (10 μg/ml) for 30 min at 4°C and washed twice with phosphate-buffered saline (PBS). Cells were then incubated with anti-human IgG Fc antibody for 20 min at 4°C. IgG-FcBright, IgG-Fcdim, and GFP− cells were purified by FACS. Cell-associated HIV-1 RNA was quantitated by the gag and vpu/env assays.
Antibody-mediated killing assay with or without patient CD4+ T cells.
CD8-depleted healthy donor PBMCs were plated in 48-well plates (0.3 to 0.5 million cells per well) and then infected with HIVivoHA at 1,200 × g at room temperature for 2 h. Cells were then washed twice with PBS to remove cell-free viruses and cultured in fresh medium containing 20 ng/ml IL-2 and 3 μg/ml anti-HA or anti-HA-GRLR antibodies. At day 4 postinfection, the percentage of HA+ Gag+ cells in CD3+ CD8− T cells of each group were measured by flow cytometry. To quantify residual HIV-1 RNA, total cellular RNA was isolated from antibody-treated or control groups. Cell-associated HIV-1 RNA was measured by the gag and vpu/env assays.
To measure ADCC, human blood NK cells were purified using the EasySep human NK cell isolation kit (catalog no. 17955; Stemcell Technologies). HIVivo-HA-infected primary CD4+ T cells were cocultured with autologous NK cells (1:1 effector to target ratio) for 6 h. Anti-HA or anti-HA-GRLR antibodies were provided at 3 μg/ml. The frequency of HA+ Gag+ 7-AAD− cells in CD3+ CD8− T cells was measured by flow cytometry.
To measure ADCP, blood monocytes were isolated using the MojoSort human CD14 selection kit (catalog no. 480026; BioLegend) and stimulated with 50 ng/ml macrophage colony-stimulating factor (M-CSF, catalog no. 574806; BioLegend) and 50 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF, catalog no. 572904; BioLegend) for 6 days. At day 6, the differentiated macrophages were treated with Accutase (catalog no. 423201; BioLegend) and replated at 0.25 to 0.5 million cells/well in a 48-well non-tissue-culture-treated plate and rested overnight before coculture with HIVivo-HA-infected autologous CD4+ T cells at a 1:1 effector to target ratio. Nonadherent cells were collected from the culture medium at 6 h post coculture. The percentage of remaining infected cells (CD14− CD3+ CD8− HA+) was determined by flow cytometry.
To test whether the vpu/env assay could be accurate when samples contained a large amount of defective HIV-1 transcripts, CD8-depleted healthy donor PBMCs were cultured and infected with HIVivoHA with or without antibody treatment as described above. At day 4 postinfection, infected cell cultures (contained <5% HA+ Gag+ cells) were diluted with untreated CD4+ T cells of HIV-1 patients on ART at a ratio of 1 to 10,000. After dilution, the frequency of HA+ Gag+ cells was reduced to less than 5 per million. The dilution was performed right before RNA extraction to avoid T-cell alloreactivity. After RNA extraction and reverse transcription, cell-associated HIV-1 RNA was quantitated by the gag and vpu/env assays.
Generation of humanized MISTRG-6-15 mice.
All animal experiments were approved by the Institutional Animal Care and Use Committee of the Washington University School of Medicine. The immunodeficient mouse strain named MISTRG-6-15 was generated by Regeneron Pharmaceuticals and the Richard Flavell laboratory at Yale University. The MISTRG-6-15 mouse carries knock-ins of human M-CSF, GM-CSF, IL-3, SIRPA, thrombopoietin (THPO), IL-6, and IL-15 coding genes on a BALB/c-Rag2−/− Il2rgnull background (50–52). To generate humanized mice, human cord blood CD34+ cells were isolated using the EasySep CD34 positive selection kit II (Stemcell Technologies). Newborn mice (1 to 3 days old) were engrafted with 20,000 cord blood CD34+ cells by intrahepatic injection. Reconstitution of human CD45+ cells in blood was determined 9 weeks postengraftment.
HIV-1 infection, antibody treatment, and viral RNA quantification in humanized mice.
At 10 weeks postengraftment, the MISTRG-6-15 mice were infected with HIV-1Ba-L (10 ng p24 per mouse) by retro-orbital injection. HIV-1 bNabs PGT121 and N6 were injected intraperitoneally at a dose of 500 μg per mouse (∼20 mg/kg) twice at day 15 and day 18 postinfection. To quantify plasma HIV-1 RNA, blood samples were collected by retro-orbital or submandibular bleeding and processed using Quick-RNA viral kits (Zymo Research) for extraction of viral RNA. After reverse transcription using SuperScript III reverse transcriptase (Thermo Fisher), HIV-1 gag qPCR was used to quantify plasma HIV-1 RNA levels. To quantify HIV-1 infection in the tissues, tissue RNA was extracted using Direct-zol RNA kits (Zymo Research). Both HIV-1 gag and vpu/env RT-qPCR assays were used to quantify tissue HIV-1 RNA.
Statistical analysis.
Statistical analyses were performed using Prism 9 (GraphPad). The methods for statistical analysis are included in the figure legends.
ACKNOWLEDGMENTS
We thank the volunteers for participating in this study. We thank Lisa Kessels, Michael Klebert, Alem Haile, Teresa Spitz, and Timira Minor for patient recruitment. We thank Regeneron Pharmaceuticals and the Richard Flavell laboratory at Yale University for generating the human cytokine knock-in mice. We thank Michel Nussenzweig for providing the HIVivoHA viral plasmid and HA antibody plasmids. We thank Dennis Burton for providing PGT121 antibody plasmids. We thank Robert Siliciano for providing pNL4-3-Δ6-drEGFP and packaging vector pC-Help plasmids. We thank the AIDS Reagent Program for providing the reagents.
This project was supported by NIAID grant R21AI150418 (to L. Shan).
REFERENCES
- 1.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291–1295. 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 2.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188. 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 3.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, Lloyd AL, Nowak MA, Fauci AS. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 94:13193–13197. 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chun TW, Finzi D, Margolick J, Chadwick K, Schwartz D, Siliciano RF. 1995. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat Med 1:1284–1290. 10.1038/nm1295-1284. [DOI] [PubMed] [Google Scholar]
- 5.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 6.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. 2009. The challenge of finding a cure for HIV infection. Science 323:1304–1307. 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 7.Shan L, Deng K, Shroff NS, Durand CM, Rabi SA, Yang HC, Zhang H, Margolick JB, Blankson JN, Siliciano RF. 2012. Stimulation of HIV-1-specific cytolytic T lymphocytes facilitates elimination of latent viral reservoir after virus reactivation. Immunity 36:491–501. 10.1016/j.immuni.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burton DR, Hangartner L. 2016. Broadly Neutralizing Antibodies to HIV and Their Role in Vaccine Design. Annu Rev Immunol 34:635–659. 10.1146/annurev-immunol-041015-055515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bar KJ, Sneller MC, Harrison LJ, Justement JS, Overton ET, Petrone ME, Salantes DB, Seamon CA, Scheinfeld B, Kwan RW, Learn GH, Proschan MA, Kreider EF, Blazkova J, Bardsley M, Refsland EW, Messer M, Clarridge KE, Tustin NB, Madden PJ, Oden K, O’Dell SJ, Jarocki B, Shiakolas AR, Tressler RL, Doria-Rose NA, Bailer RT, Ledgerwood JE, Capparelli EV, Lynch RM, Graham BS, Moir S, Koup RA, Mascola JR, Hoxie JA, Fauci AS, Tebas P, Chun T-W. 2016. Effect of HIV antibody VRC01 on viral rebound after treatment interruption. N Engl J Med 375:2037–2050. 10.1056/NEJMoa1608243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schoofs T, Klein F, Braunschweig M, Kreider EF, Feldmann A, Nogueira L, Oliveira T, Lorenzi JC, Parrish EH, Learn GH, West AP, Jr, Bjorkman PJ, Schlesinger SJ, Seaman MS, Czartoski J, McElrath MJ, Pfeifer N, Hahn BH, Caskey M, Nussenzweig MC. 2016. HIV-1 therapy with monoclonal antibody 3BNC117 elicits host immune responses against HIV-1. Science 352:997–1001. 10.1126/science.aaf0972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caskey M, Schoofs T, Gruell H, Settler A, Karagounis T, Kreider EF, Murrell B, Pfeifer N, Nogueira L, Oliveira TY, Learn GH, Cohen YZ, Lehmann C, Gillor D, Shimeliovich I, Unson-O'Brien C, Weiland D, Robles A, Kummerle T, Wyen C, Levin R, Witmer-Pack M, Eren K, Ignacio C, Kiss S, West AP, Jr, Mouquet H, Zingman BS, Gulick RM, Keler T, Bjorkman PJ, Seaman MS, Hahn BH, Fatkenheuer G, Schlesinger SJ, Nussenzweig MC, Klein F. 2017. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat Med 23:185–191. 10.1038/nm.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Scheid JF, Horwitz JA, Bar-On Y, Kreider EF, Lu CL, Lorenzi JC, Feldmann A, Braunschweig M, Nogueira L, Oliveira T, Shimeliovich I, Patel R, Burke L, Cohen YZ, Hadrigan S, Settler A, Witmer-Pack M, West AP, Jr, Juelg B, Keler T, Hawthorne T, Zingman B, Gulick RM, Pfeifer N, Learn GH, Seaman MS, Bjorkman PJ, Klein F, Schlesinger SJ, Walker BD, Hahn BH, Nussenzweig MC, Caskey M. 2016. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature 535:556–560. 10.1038/nature18929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu CL, Murakowski DK, Bournazos S, Schoofs T, Sarkar D, Halper-Stromberg A, Horwitz JA, Nogueira L, Golijanin J, Gazumyan A, Ravetch JV, Caskey M, Chakraborty AK, Nussenzweig MC. 2016. Enhanced clearance of HIV-1-infected cells by broadly neutralizing antibodies against HIV-1 in vivo. Science 352:1001–1004. 10.1126/science.aaf1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendoza P, Gruell H, Nogueira L, Pai JA, Butler AL, Millard K, Lehmann C, Suarez I, Oliveira TY, Lorenzi JCC, Cohen YZ, Wyen C, Kummerle T, Karagounis T, Lu CL, Handl L, Unson-O'Brien C, Patel R, Ruping C, Schlotz M, Witmer-Pack M, Shimeliovich I, Kremer G, Thomas E, Seaton KE, Horowitz J, West AP, Jr, Bjorkman PJ, Tomaras GD, Gulick RM, Pfeifer N, Fatkenheuer G, Seaman MS, Klein F, Caskey M, Nussenzweig MC. 2018. Combination therapy with anti-HIV-1 antibodies maintains viral suppression. Nature 561:479–484. 10.1038/s41586-018-0531-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC, Richman DD, Hudgens MG, Bosch RJ, Coffin JM, Eron JJ, Hazuda DJ, Margolis DM. 2012. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature 487:482–485. 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott JH, Wightman F, Solomon A, Ghneim K, Ahlers J, Cameron MJ, Smith MZ, Spelman T, McMahon J, Velayudham P, Brown G, Roney J, Watson J, Prince MH, Hoy JF, Chomont N, Fromentin R, Procopio FA, Zeidan J, Palmer S, Odevall L, Johnstone RW, Martin BP, Sinclair E, Deeks SG, Hazuda DJ, Cameron PU, Sekaly RP, Lewin SR. 2014. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog 10:e1004473. 10.1371/journal.ppat.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cillo AR, Sobolewski MD, Bosch RJ, Fyne E, Piatak M, Jr, Coffin JM, Mellors JW. 2014. Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A 111:7078–7083. 10.1073/pnas.1402873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. 2014. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med 20:425–429. 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho YC, Richman DD, Deeks SG, Siliciano JD, Siliciano RF. 2016. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 22:1043–1049. 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, Bertagnolli LN, Capoferri AA, Kufera JT, Timmons A, Nobles C, Gregg J, Wada N, Ho YC, Zhang H, Margolick JB, Blankson JN, Deeks SG, Bushman FD, Siliciano JD, Laird GM, Siliciano RF. 2019. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566:120–125. 10.1038/s41586-019-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yukl SA, Kaiser P, Kim P, Telwatte S, Joshi SK, Vu M, Lampiris H, Wong JK. 2018. HIV latency in isolated patient CD4+ T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci Transl Med 10. 10.1126/scitranslmed.aap9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasternak AO, Adema KW, Bakker M, Jurriaans S, Berkhout B, Cornelissen M, Lukashov VV. 2008. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J Clin Microbiol 46:2206–2211. 10.1128/JCM.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Procopio FA, Fromentin R, Kulpa DA, Brehm JH, Bebin AG, Strain MC, Richman DD, O'Doherty U, Palmer S, Hecht FM, Hoh R, Barnard RJ, Miller MD, Hazuda DJ, Deeks SG, Sekaly RP, Chomont N. 2015. A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine 2:874–883. 10.1016/j.ebiom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das B, Dobrowolski C, Luttge B, Valadkhan S, Chomont N, Johnston R, Bacchetti P, Hoh R, Gandhi M, Deeks SG, Scully E, Karn J. 2018. Estrogen receptor-1 is a key regulator of HIV-1 latency that imparts gender-specific restrictions on the latent reservoir. Proc Natl Acad Sci U S A 115:E7795–E7804. 10.1073/pnas.1803468115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartz S, Felber BK, Fenyo EM, Pavlakis GN. 1990. Env and Vpu proteins of human immunodeficiency virus type 1 are produced from multiple bicistronic mRNAs. J Virol 64:5448–5456. 10.1128/JVI.64.11.5448-5456.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartz S, Felber BK, Pavlakis GN. 1992. Mechanism of translation of monocistronic and multicistronic human immunodeficiency virus type 1 mRNAs. Mol Cell Biol 12:207–219. 10.1128/mcb.12.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiener B, Horsburgh BA, Eden JS, Barton K, Schlub TE, Lee E, von Stockenstrom S, Odevall L, Milush JM, Liegler T, Sinclair E, Hoh R, Boritz EA, Douek D, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S. 2017. Identification of genetically intact HIV-1 proviruses in specific CD4+ T cells from effectively treated participants. Cell Rep 21:813–822. 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ocwieja KE, Sherrill-Mix S, Mukherjee R, Custers-Allen R, David P, Brown M, Wang S, Link DR, Olson J, Travers K, Schadt E, Bushman FD. 2012. Dynamic regulation of HIV-1 mRNA populations analyzed by single-molecule enrichment and long-read sequencing. Nucleic Acids Res 40:10345–10355. 10.1093/nar/gks753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim Y, Anderson JL, Lewin SR. 2018. Getting the “kill” into “shock and kill”: strategies to eliminate latent HIV. Cell Host Microbe 23:14–26. 10.1016/j.chom.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deeks SG. 2012. HIV: shock and kill. Nature 487:439–440. 10.1038/487439a. [DOI] [PubMed] [Google Scholar]
- 32.Cary DC, Peterlin BM. 2016. Targeting the latent reservoir to achieve functional HIV cure. F1000Res 5:1009. 10.12688/f1000research.8109.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang HC, Xing S, Shan L, O'Connell K, Dinoso J, Shen A, Zhou Y, Shrum CK, Han Y, Liu JO, Zhang H, Margolick JB, Siliciano RF. 2009. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J Clin Invest 119:3473–3486. 10.1172/JCI39199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xing S, Bullen CK, Shroff NS, Shan L, Yang HC, Manucci JL, Bhat S, Zhang H, Margolick JB, Quinn TC, Margolis DM, Siliciano JD, Siliciano RF. 2011. Disulfiram reactivates latent HIV-1 in a Bcl-2-transduced primary CD4+ T cell model without inducing global T cell activation. J Virol 85:6060–6064. 10.1128/JVI.02033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han Y, Lassen K, Monie D, Sedaghat AR, Shimoji S, Liu X, Pierson TC, Margolick JB, Siliciano RF, Siliciano JD. 2004. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J Virol 78:6122–6133. 10.1128/JVI.78.12.6122-6133.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521–529. 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 37.Sogaard OS, Graversen ME, Leth S, Olesen R, Brinkmann CR, Nissen SK, Kjaer AS, Schleimann MH, Denton PW, Hey-Cunningham WJ, Koelsch KK, Pantaleo G, Krogsgaard K, Sommerfelt M, Fromentin R, Chomont N, Rasmussen TA, Ostergaard L, Tolstrup M. 2015. The depsipeptide romidepsin reverses HIV-1 latency in vivo. PLoS Pathog 11:e1005142. 10.1371/journal.ppat.1005142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruelas DS, Greene WC. 2013. An integrated overview of HIV-1 latency. Cell 155:519–529. 10.1016/j.cell.2013.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geeraert L, Kraus G, Pomerantz RJ. 2008. Hide-and-seek: the challenge of viral persistence in HIV-1 infection. Annu Rev Med 59:487–501. 10.1146/annurev.med.59.062806.123001. [DOI] [PubMed] [Google Scholar]
- 40.Turner AW, Margolis DM. 2017. Chromatin regulation and the histone code in HIV latency. Yale J Biol Med 90:229–243. [PMC free article] [PubMed] [Google Scholar]
- 41.Margolis DM, Archin NM, Cohen MS, Eron JJ, Ferrari G, Garcia JV, Gay CL, Goonetilleke N, Joseph SB, Swanstrom R, Turner AW, Wahl A. 2020. Curing HIV: seeking to target and clear persistent infection. Cell 181:189–206. 10.1016/j.cell.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Overbaugh J, Morris L. 2012. The antibody response against HIV-1. Cold Spring Harb Perspect Med 2:a007039. 10.1101/cshperspect.a007039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horwitz JA, Bar-On Y, Lu CL, Fera D, Lockhart AAK, Lorenzi JCC, Nogueira L, Golijanin J, Scheid JF, Seaman MS, Gazumyan A, Zolla-Pazner S, Nussenzweig MC. 2017. Non-neutralizing antibodies alter the course of HIV-1 infection in vivo. Cell 170:637–648.e10. 10.1016/j.cell.2017.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sato K, Izumi T, Misawa N, Kobayashi T, Yamashita Y, Ohmichi M, Ito M, Takaori-Kondo A, Koyanagi Y. 2010. Remarkable lethal G-to-A mutations in vif-proficient HIV-1 provirus by individual APOBEC3 proteins in humanized mice. J Virol 84:9546–9556. 10.1128/JVI.00823-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, Zang T, Dorner M, Billerbeck E, Labitt RN, Gaebler C, Marcovecchio P, Incesu RB, Eisenreich TR, Bieniasz PD, Seaman MS, Bjorkman PJ, Ravetch JV, Ploss A, Nussenzweig MC. 2012. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature 492:118–122. 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Halper-Stromberg A, Lu CL, Klein F, Horwitz JA, Bournazos S, Nogueira L, Eisenreich TR, Liu C, Gazumyan A, Schaefer U, Furze RC, Seaman MS, Prinjha R, Tarakhovsky A, Ravetch JV, Nussenzweig MC. 2014. Broadly neutralizing antibodies and viral inducers decrease rebound from HIV-1 latent reservoirs in humanized mice. Cell 158:989–999. 10.1016/j.cell.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borducchi EN, Liu J, Nkolola JP, Cadena AM, Yu WH, Fischinger S, Broge T, Abbink P, Mercado NB, Chandrashekar A, Jetton D, Peter L, McMahan K, Moseley ET, Bekerman E, Hesselgesser J, Li W, Lewis MG, Alter G, Geleziunas R, Barouch DH. 2018. Publisher correction: Antibody and TLR7 agonist delay viral rebound in SHIV-infected monkeys. Nature 564:E8. 10.1038/s41586-018-0721-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laird GM, Eisele EE, Rabi SA, Lai J, Chioma S, Blankson JN, Siliciano JD, Siliciano RF. 2013. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog 9:e1003398. 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siliciano JD, Siliciano RF. 2005. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol 304:3–15. 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 50.Rongvaux A, Willinger T, Martinek J, Strowig T, Gearty SV, Teichmann LL, Saito Y, Marches F, Halene S, Palucka AK, Manz MG, Flavell RA. 2014. Development and function of human innate immune cells in a humanized mouse model. Nat Biotechnol 32:364–372. 10.1038/nbt.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Herndler-Brandstetter D, Shan L, Yao Y, Stecher C, Plajer V, Lietzenmayer M, Strowig T, de Zoete MR, Palm NW, Chen J, Blish CA, Frleta D, Gurer C, Macdonald LE, Murphy AJ, Yancopoulos GD, Montgomery RR, Flavell RA. 2017. Humanized mouse model supports development, function, and tissue residency of human natural killer cells. Proc Natl Acad Sci U S A 114:E9626–E9634. 10.1073/pnas.1705301114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu H, Borsotti C, Schickel JN, Zhu S, Strowig T, Eynon EE, Frleta D, Gurer C, Murphy AJ, Yancopoulos GD, Meffre E, Manz MG, Flavell RA. 2017. A novel humanized mouse model with significant improvement of class-switched, antigen-specific antibody production. Blood 129:959–969. 10.1182/blood-2016-04-709584. [DOI] [PMC free article] [PubMed] [Google Scholar]