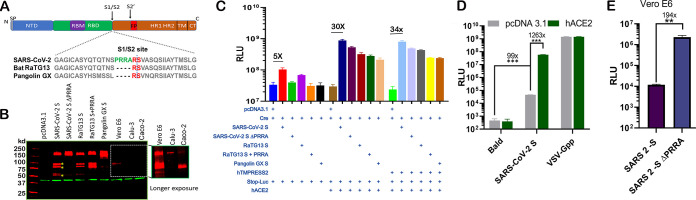
FIG 2.
PRRA-led proteolytic cleavage of SARS-CoV-2 spike protein and effect on fusion. (A) Organization of SARS-CoV-2 spike protein and sequence alignment at the S1/S2 site with RaTG13 and pangolin GX S proteins. SP, signal peptide; NTD, N-terminal domain; FP, fusion peptide; TM, transmembrane; CT, C terminus. (B) 293T cells transfected with SARS-CoV-2 S, SARS-CoV-2 S ΔPRRA, bat RaTG13 spike (RaTG13 S), an insertion mutant containing PRRA (RaTG13 S+PRRA), and the pangolin GX spike protein (Pangolin GX S) (lanes 3 to 7) and SARS-CoV-2 S-infected Vero E6, Calu-3, and Caco-2 cells (lanes 8 to 10). A 5-min exposure of this part of the gel is included. Red, anti-S antibody; green, anti-β-actin antibody. (C) Cell-cell fusion mediated by CoV S proteins. 293T cells expressing Stop‐Luc, ACE2, and/or ACE2/TMPRSS2 (acceptor cells) were mixed at a 1:1 ratio with donor cells expressing Cre, CoV S, or both to initiate cell‐cell fusion. Data are presented as means ± standard errors of the means (SEM). (D) 293T cells or 293T-hACE2 cells were infected by pseudovirus bearing SARS-CoV-2 S. Bald viruses without any viral envelope or VSV-Gpp were included as negative and positive controls. (E) Entry of MLV pseudoviruses bearing SARS-CoV-2 S or SARS-CoV-2 S ΔPRRA in Vero cells. **, P < 0.001; ***, P < 0.0001. RLU, relative luciferase units.