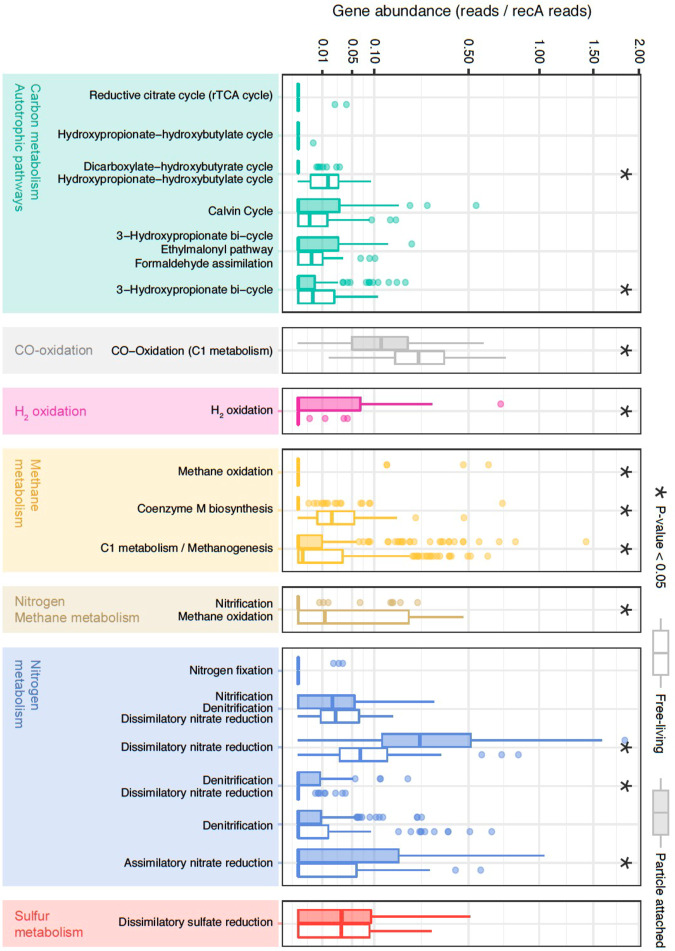
Fig. 4. Comparison of metabolic pathways in the free-living and particle-attached microbial communities from the bathypelagic ocean.
Normalized gene abundance (reads/recA reads) of 49 marker genes indicative of different autotrophic carbon-fixation pathways, nitrogen, sulfur, methane, and hydrogen metabolisms in the Malaspina Gene DataBase. Gene abundances from KOs belonging to the same pathway and KEGG module level have been plotted together (Supplementary Data 7). From top to the bottom: Reductive citrate cycle (rTCA): K15230, K15231); hydroxypropionate–hydroxybutyrate cycle: K15039; dicarboxylate-hydroxybutyrate cycle|hydroxypropionate-hydroxybutylate cycle: K15016; Calvin cycle: K00855; K01601 and K01602; 3-hydroxypropionate bi-cycle|ethylmalonyl pathway|formaldehyde assimilation: K08691; 3-hydroxypropionate bi-cycle: K14468, K14470 and K09709; CO oxidation (C1 metabolism): K03520; K03519 and K03518; H2-oxidation: K00436; methane oxidation: K16158; coenzyme M biosynthesis: K05979, K06034, K08097 and K13039; C1 metabolism/methanogenesis: K00320, K00200, K00201, K00202, K00672, K03390, K14083, and K00577; nitrification|methane oxidation: K10944, K10945, and K10946; Nitrogen fixation: K02588; nitrification|denitrification|dissimilatory nitrate reduction: K00370 and K00371; dissimilatory nitrate reduction (DNRA): K00362 and K00363; denitrification|dissimilatory nitrate reduction: K00374, K02567, and K02568; denitrification: K00368, K15864, K04561, K02305, and K00376; assimilatory nitrate reduction: K00367 and K00372; dissimilatory sulfate reduction: K00394, K00395 and K11181. Wilcoxon tests were done to test for significant differences between the particle-attached (PA) and free-living (FL) assemblages and significant (P value < 0.05) differences are labeled with asterisks. FL (empty boxes) and PA (filled boxes) bathypelagic microbial communities are shown next to each other.