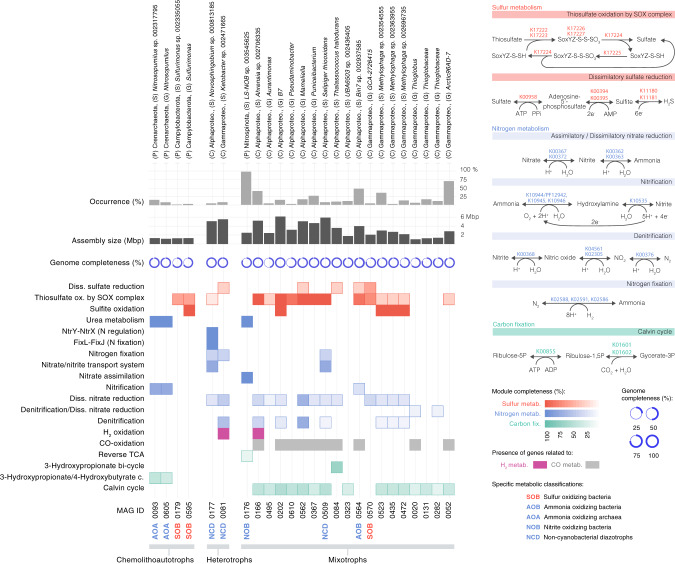
Fig. 6. Metabolic potential in selected metagenome-assembled genomes (MAGs) from the bathypelagic ocean.
A total of 25 MDeep-MAGs were selected based on the presence of metabolic pathways with potential for chemolithoautotrophy, mixotrophy, and nitrogen fixation (non-cyanobacterial diazotrophs, NCDs). AOA ammonia-oxidizing archaea, SOB sulfur-oxidizing bacteria, NCDs non-cyanobacterial diazotrophs, NOB nitrite-oxidizing bacteria, AOB ammonia-oxidizing bacteria. Their taxonomic assignment at the maximal possible resolution is shown at the top, followed by the occurrence of each MAG in samples of the Malaspina Gene DataBase, and the genome completeness (%) of each MAG. On the right, metabolic pathways involved in inorganic carbon fixation (green), sulfur (red), nitrogen (blue) are shown as well as the KOs that participate in these pathways. At the bottom, the percentage module completeness for each pathway is coded by color intensity.