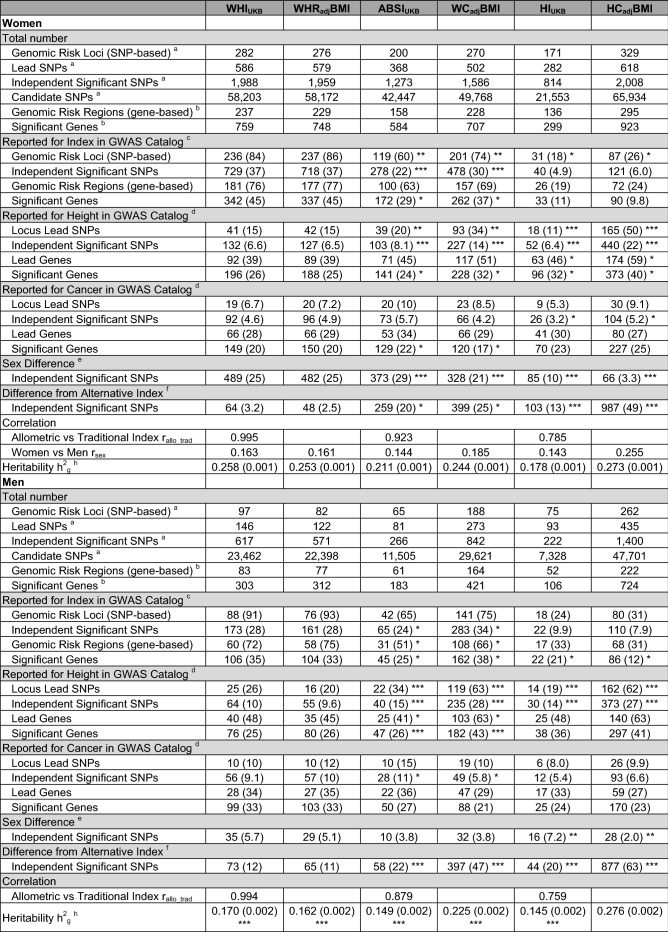
Table 1.
Counts and overlaps of independent genetic variants and loci associated with allometric and traditional body-shape indices in women and men.
aBased on FUMA; bBased on MAGMA; cNumber (percentage from total per index) reported in the NHGRI-EBI GWAS Catalog11 (https://www.ebi.ac.uk/gwas/home, accessed on 07/04/2021) in association with the corresponding traditional body-shape index (with or without adjustment for body mass index, BMI), i.e. the waist-to-hip ratio for WHIUKB and WHRadjBMI (catalogue sets EFO_0004343, EFO_0007788, EFO_0004302), waist circumference for ABSIUKB and WCadjBMI (EFO_0004342, EFO_0007789, EFO_0004302), hip circumference for HIUKB and HCadjBMI (EFO_0005093, EFO_0008039, EFO_0004302). To ensure novelty, a match was counted for an independent significant SNP if any of the candidate SNPs in the LD block was reported and for a genomic risk locus (or region) if any of the corresponding independent significant SNPs (or genes) was reported; dNumber (percentage from total per index), counting indirect matches only for SNPs in strong LD, i.e. candidate SNPs within the corresponding LD block for independent significant SNPs and for a genomic risk locus (or region) only a match of the independent significant SNP (or gene) promoted to a locus lead SNP (or lead gene) (EFO_0004339, EFO_0004302 for height; EFO_0000311 for cancer); eNumber (percentage from total per index) showing sex difference (pdifference < 5*10–6 for any candidate SNP within the corresponding LD block); fAs for ebut reflecting difference from the corresponding alternative body-shape index, i.e. WHRadjBMI for WHIUKB, WCadjBMI for ABSIUKB, HCadjBMI for HIUKB and vice versa; ABSIUKB-a body shape index calibrated for UK Biobank participants; HCadjBMI-hip circumference adjusted for BMI; HIUKB-hip index calibrated for UK Biobank participants; WCadjBMI-waist circumference adjusted for BMI; WHIUKB-waist-to-hip index calibrated for UK Biobank participants; WHRadjBMI-waist-to-hip ratio adjusted BMI; h2g-estimated (pseudo-) heritability, based on the genetic relationship matrix in BOLT-LMM (approximate standard error, 316/number of individuals), comparison between men and women; rsex-Spearman’s rank correlation coefficient between regression coefficients in women and men across all examined genetic variants (used in the test for difference of effect size or heritability); rallo-trad-correlation coefficient as for rsex but between the corresponding allometric and traditional index; Fisher’s exact test was used to compare percentages between allometric and traditional body-shape indices: *P < 0.05; **P < 0.001; ***P < 0.0001.