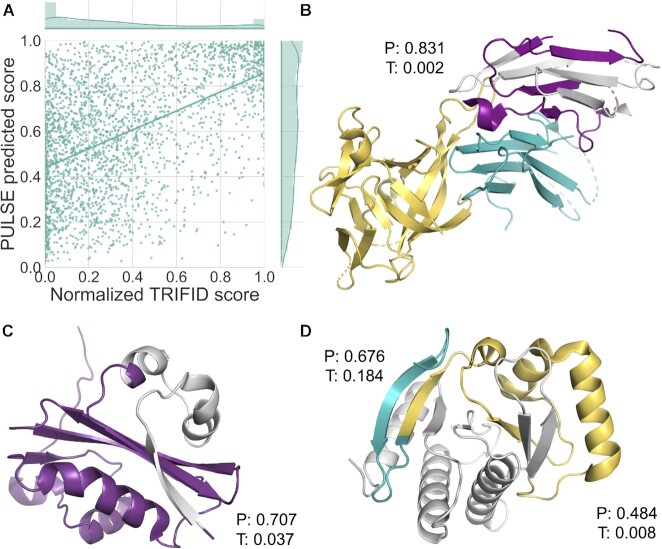
Figure 7.
A comparison between TRIFID and PULSE. (A) A scatter plot of PULSE and TRIFID scores over alternative isoforms that coincide between the two analyses. The comparison was carried out over 2692 sequences present in both data sets. The distribution of scores for the predictors is shown above or to the right of the graphic. Spearman's rank correlation between the two sets was 0.504. (B) The 346-residue splice variant of IL1RAP mapped onto PDB structure 5VI4. This isoform is generated from an exon skip that changes the frame of the protein. The exon skip occurs in the middle of the third immunoglobulin domain (in purple) and as a result of the frame shift, the variant loses half of the domain (lost region shown in light grey) and the downstream trans-membrane helix and downstream TIR domain. The interaction with interleukin-33 (yellow) and interleukin 1 receptor like 1 (teal) will also be affected. The isoform is annotated only in the human genome. PULSE predicts that this isoform is functional (0.831), while TRIFID does not (0.002). (C) The 475-residue splice variant of ATE1 mapped onto PDB structure 2ATR using HHPRED. This isoform is generated from an exon skip that removes 41 residues including the first part of the Arginine-tRNA-protein transferase domain (lost region shown in light grey). This splice event skips a pair of mutually exclusively spliced exons that appear to be important in substrate selection (85) and that are conserved even in Orb weaver spiders. It seems unlikely that such important exons can be skipped without consequence for the function of the protein. PULSE predicts that this isoform is functional (0.707) and TRIFID does not (0.037). (D) Two splice variants of MACROH2A1 mapped onto PDB structure 6fy5 using HHPRED. The first isoform is generated from an exon skip that changes the frame at the start of the macro domain. The section of the structure that would be maintained is shown in teal, the remainder (in yellow and light grey) would be replaced by 27 residues as a result of the frame shift. PULSE predicts that this isoform is functional (0.676), while TRIFID does not (0.184). A second exon skip produces another frame shift that affects the same domain. Here the conserved region is shown in purple and yellow, and the region of the domain replaced by frame-shifted residues in light grey. Neither method predicts that this isoform is functional, but the PULSE score for this improbable protein is much higher, 0.484 against 0.008. All images were generated using PyMol.