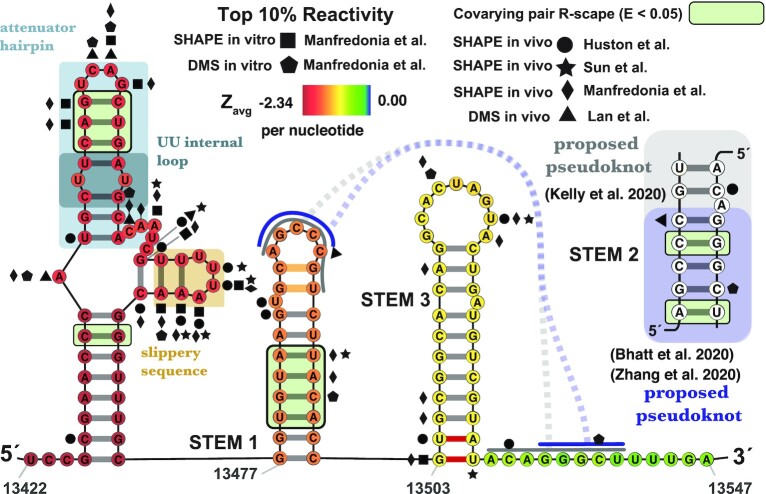
Figure 1.
In silico ScanFold-Fold predicted secondary structure for the SARS-CoV-2 frameshift stimulatory element (FSE) spanning nts 13422–13547. Average z-scores are overlaid on each nt via a heat map ranging from –2.34 (red) to 0.00 (blue). Top 10% of reactivities are shown for Manfredoina et al. (squares, pentagons and diamonds), Huston et al. (circles), Sun et al. (stars) and Lan et al. (triangles) at their corresponding nt positions (17–20). The attenuator hairpin and UU internal loop, recently targeted with small molecule inhibitors of –1 PRF (16), are depicted in blue shaded boxes and the slippery sequence in a gold shaded box. The interactions of the pseudoknot proposed by other groups (17–20) are shown with solid and dashed gray lines and the specific base pairing pattering are also shown in an inset. The smaller pseudoknot structure as determined by cyro-EM (14,73) is highlighted in lavender (dashed line and inset). The two orange colored pairs at the top of Stem 1 were not detected by Bhatt et al. (73) in their cryo-EM and the two red pairs at the base of stem 3 were not detected by either Bhatt et al. or Zhang et al. (14,73). All significantly covarying bps (R-scape APC corrected G-test; E < 0.05) have been highlighted with a green box.