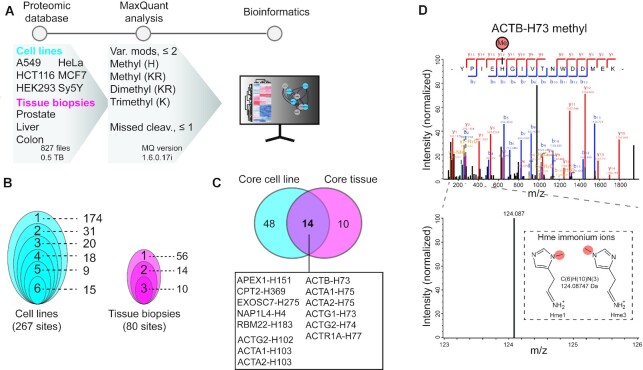
Figure 2.
Exploring histidine methylation in human cells and tissues. (A) Methylome profiling workflow. Publicly available ultra-comprehensive proteomics datasets from human cell lines and tissue biopsies were searched for histidine, lysine and arginine methylation using MaxQuant. Identified histidine methylation events were analyzed to explore the abundance and cellular context of the PTM. (B) Clam plot representation of histidine methylation sites identified in cells and tissues. The total number of sites and the number of shared sites between cell lines (left) and tissue biopsies (right) are indicated. (C) Core histidine methylation sites. Sites were categorized as part of the core methylome if identified in more than 50% of the samples in each category. (D) Mass spectra supporting methylation of ACTB-H73. Tandem mass for a tryptic peptide covering Y69-K84 of ACTB unambiguously supporting methylation of H73. The presence of a specific immonium ion corresponding to methyl-histidine is indicated.