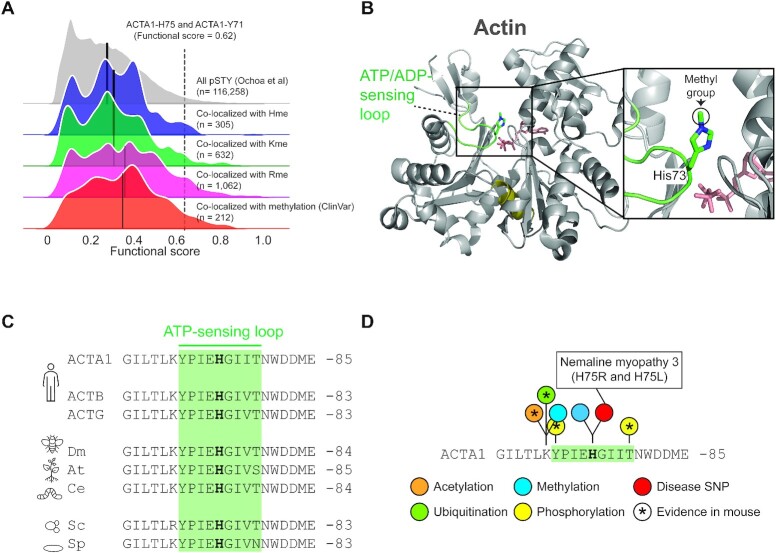
Figure 5.
PTM colocalization. (A) Kernel density plots for the functional score distributions of colocalizing phosphorylation sites with identified methylation events. Subsets of phosphorylation sites colocalizing with Rme, Kme or Hme methylation sites plotted separately. Separate grouping of phosphorylation sites co-localizing with methylation sites with a reported polymorphism or mutation associated with a pathological condition (ClinVar) is shown. Black line indicate group mean. Methylation of ACTA1-H75 and the colocalizing phosphorylation event on ACTA1-Y71 are indicated. (B) The structural context of actin histidine methylation. The structure of actin is shown in cartoon representation whereas ATP and the methylated histidine residue H73 is shown in stick representation. The hinge region (olive), ATP (salmon) and the H73 containing ATP-sensing loop (green) are indicated. The structure is derived from rabbit muscle alpha actin (PDB #1EQYE). (C) Evolutionary conservation of the methyl-histidine containing loop in actin. Sequences: human ACTA1 (P68133), ACTB (P60709) and ACTG (P63261) as well as homologues from Drosophila melanogaster (dm; AAA28314.1), Arabidopsis thaliana (at; NP_187818.1), Caenorhabditis elegans (ce; NP_508841.1), Saccharomyces cerevisiae (cs; NP_116614.1) and Saccharomyces pombe (sp; NP_595618.1). (D) The methyl-histidine containing loop in ACTA1 is a PTM hotspot. PTMs annotated in the PhosphoSitePlus database (v6.5.9.3) are shown. Modifications observed in mouse ACTA1 (star) and sites corresponding to disease associate mutations are indicated (red).